
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,945,131 – 20,945,291 |
| Length | 160 |
| Max. P | 0.983612 |

| Location | 20,945,131 – 20,945,251 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -38.65 |
| Energy contribution | -38.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.65 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20945131 120 + 22407834 CUCCUCCUCCAGCGACCUAGAGUUUCCGAAAAGUUCCCACUUCGGUUUCAAAUUCAGAUUGAGAGCCAGAUUCAGAGCCAGUGGAGUUUGCUUUUUCUGGCCUGGUCUCUGGCAUUGAGG .....((((...(((.((.((((((((((..(((....)))))))....)))))))).)))...((((((.((((.(((((.((((....))))..)))))))))..))))))...)))) ( -39.20) >DroSec_CAF1 31346 120 + 1 CUCCUCCUCCAGCGACCUAGAGUUUUCGAAAAGUUCCCACUUCGGUUUCAAAUUCAGAUUGAGAGCCAGAUUCAGAGCCAGUGGAGUUUGCUUUUUCUGGCCUGGUCUCUGGCAUUGAGG .....((((...(((.((.((((((((((..(((....)))))))....)))))))).)))...((((((.((((.(((((.((((....))))..)))))))))..))))))...)))) ( -36.90) >DroSim_CAF1 28417 120 + 1 CUCCUCCUCCAGCGACCUAGAGUUUCCGAAAAGUUCCCACUUCGGUUUCAAAUUCAGAUUGAGAGCCAGAUUCAGAGCCAGUGGAGUUUGCUUUUUCUGGCCUGGUCUCUGGCAUUGAGG .....((((...(((.((.((((((((((..(((....)))))))....)))))))).)))...((((((.((((.(((((.((((....))))..)))))))))..))))))...)))) ( -39.20) >consensus CUCCUCCUCCAGCGACCUAGAGUUUCCGAAAAGUUCCCACUUCGGUUUCAAAUUCAGAUUGAGAGCCAGAUUCAGAGCCAGUGGAGUUUGCUUUUUCUGGCCUGGUCUCUGGCAUUGAGG .....((((...(((.((.((((((((((..(((....)))))))....)))))))).)))...((((((.((((.(((((.((((....))))..)))))))))..))))))...)))) (-38.65 = -38.43 + -0.22)

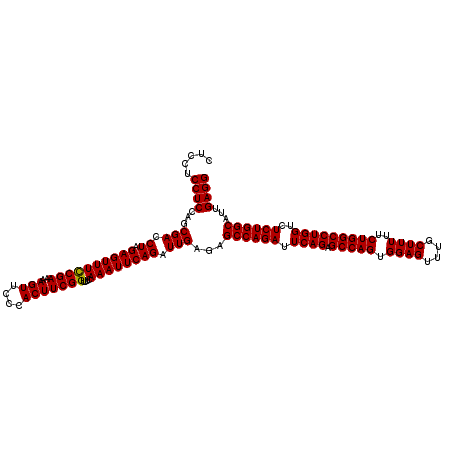
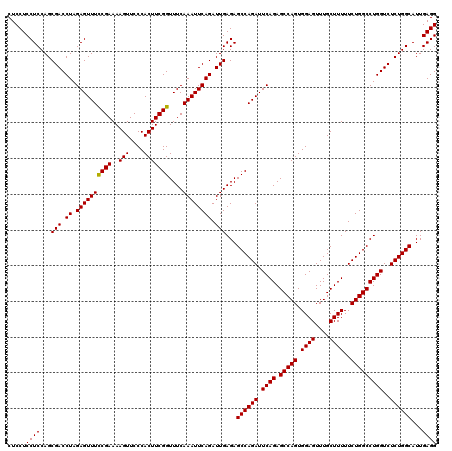
| Location | 20,945,131 – 20,945,251 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -41.17 |
| Energy contribution | -41.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20945131 120 - 22407834 CCUCAAUGCCAGAGACCAGGCCAGAAAAAGCAAACUCCACUGGCUCUGAAUCUGGCUCUCAAUCUGAAUUUGAAACCGAAGUGGGAACUUUUCGGAAACUCUAGGUCGCUGGAGGAGGAG ((((...((((((...((((((((....((....))...))))).)))..))))))..((((.......))))..((((((.(....).))))))...((((((....)))))))))).. ( -43.30) >DroSec_CAF1 31346 120 - 1 CCUCAAUGCCAGAGACCAGGCCAGAAAAAGCAAACUCCACUGGCUCUGAAUCUGGCUCUCAAUCUGAAUUUGAAACCGAAGUGGGAACUUUUCGAAAACUCUAGGUCGCUGGAGGAGGAG ((((...((((((...((((((((....((....))...))))).)))..))))))..((((.......))))...(((((.(....).)))))....((((((....)))))))))).. ( -39.80) >DroSim_CAF1 28417 120 - 1 CCUCAAUGCCAGAGACCAGGCCAGAAAAAGCAAACUCCACUGGCUCUGAAUCUGGCUCUCAAUCUGAAUUUGAAACCGAAGUGGGAACUUUUCGGAAACUCUAGGUCGCUGGAGGAGGAG ((((...((((((...((((((((....((....))...))))).)))..))))))..((((.......))))..((((((.(....).))))))...((((((....)))))))))).. ( -43.30) >consensus CCUCAAUGCCAGAGACCAGGCCAGAAAAAGCAAACUCCACUGGCUCUGAAUCUGGCUCUCAAUCUGAAUUUGAAACCGAAGUGGGAACUUUUCGGAAACUCUAGGUCGCUGGAGGAGGAG ((((...((((((...((((((((....((....))...))))).)))..))))))..((((.......))))..((((((.(....).))))))...((((((....)))))))))).. (-41.17 = -41.50 + 0.33)

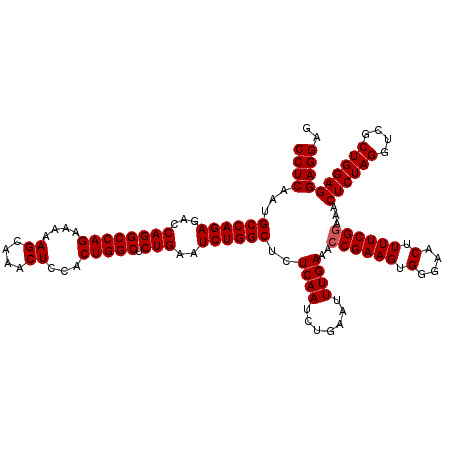
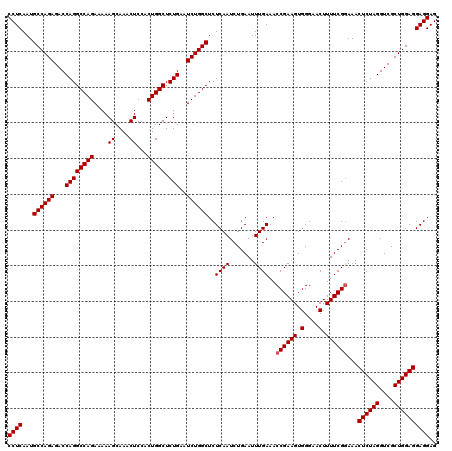
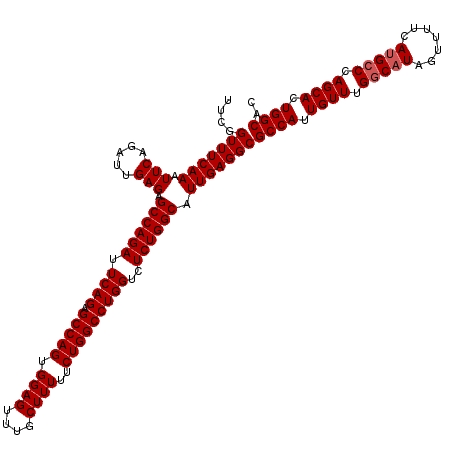
| Location | 20,945,171 – 20,945,291 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -48.23 |
| Consensus MFE | -46.63 |
| Energy contribution | -47.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20945171 120 + 22407834 UUCGGUUUCAAAUUCAGAUUGAGAGCCAGAUUCAGAGCCAGUGGAGUUUGCUUUUUCUGGCCUGGUCUCUGGCAUUGAGGCGCCAUUGUUUGGCAUAGUUUUCAAGCCCAGCACUGGCAC ....(((((((.(((.....))).((((((.((((.(((((.((((....))))..)))))))))..)))))).)))))))((((.((((.(((...........))).)))).)))).. ( -47.90) >DroSec_CAF1 31386 120 + 1 UUCGGUUUCAAAUUCAGAUUGAGAGCCAGAUUCAGAGCCAGUGGAGUUUGCUUUUUCUGGCCUGGUCUCUGGCAUUGAGGCGCCAUUGUUUGGCAUAGUUUUCAUGACCAGCACUGGCAC ....(((((((.(((.....))).((((((.((((.(((((.((((....))))..)))))))))..)))))).)))))))((((.((((.(((((.......))).)))))).)))).. ( -46.20) >DroSim_CAF1 28457 120 + 1 UUCGGUUUCAAAUUCAGAUUGAGAGCCAGAUUCAGAGCCAGUGGAGUUUGCUUUUUCUGGCCUGGUCUCUGGCAUUGAGGCGCCAUUGUUUGGCAUAGUUUUCAUGCCCAGCACUGGCAC ....(((((((.(((.....))).((((((.((((.(((((.((((....))))..)))))))))..)))))).)))))))((((.((((.(((((.......))))).)))).)))).. ( -50.60) >consensus UUCGGUUUCAAAUUCAGAUUGAGAGCCAGAUUCAGAGCCAGUGGAGUUUGCUUUUUCUGGCCUGGUCUCUGGCAUUGAGGCGCCAUUGUUUGGCAUAGUUUUCAUGCCCAGCACUGGCAC ....(((((((.(((.....))).((((((.((((.(((((.((((....))))..)))))))))..)))))).)))))))((((.((((.(((((.......))))).)))).)))).. (-46.63 = -47.30 + 0.67)


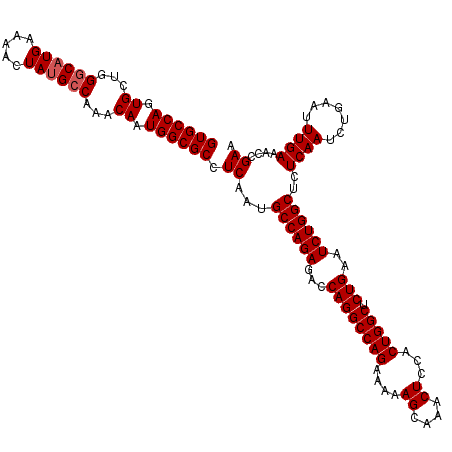
| Location | 20,945,171 – 20,945,291 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -37.43 |
| Energy contribution | -38.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20945171 120 - 22407834 GUGCCAGUGCUGGGCUUGAAAACUAUGCCAAACAAUGGCGCCUCAAUGCCAGAGACCAGGCCAGAAAAAGCAAACUCCACUGGCUCUGAAUCUGGCUCUCAAUCUGAAUUUGAAACCGAA ((((((.((...(((...........)))...)).)))))).((...((((((...((((((((....((....))...))))).)))..))))))..((((.......))))....)). ( -37.70) >DroSec_CAF1 31386 120 - 1 GUGCCAGUGCUGGUCAUGAAAACUAUGCCAAACAAUGGCGCCUCAAUGCCAGAGACCAGGCCAGAAAAAGCAAACUCCACUGGCUCUGAAUCUGGCUCUCAAUCUGAAUUUGAAACCGAA ((((((.((.(((.((((.....)))))))..)).)))))).((...((((((...((((((((....((....))...))))).)))..))))))..((((.......))))....)). ( -37.50) >DroSim_CAF1 28457 120 - 1 GUGCCAGUGCUGGGCAUGAAAACUAUGCCAAACAAUGGCGCCUCAAUGCCAGAGACCAGGCCAGAAAAAGCAAACUCCACUGGCUCUGAAUCUGGCUCUCAAUCUGAAUUUGAAACCGAA ((((((.((...((((((.....))))))...)).)))))).((...((((((...((((((((....((....))...))))).)))..))))))..((((.......))))....)). ( -41.40) >consensus GUGCCAGUGCUGGGCAUGAAAACUAUGCCAAACAAUGGCGCCUCAAUGCCAGAGACCAGGCCAGAAAAAGCAAACUCCACUGGCUCUGAAUCUGGCUCUCAAUCUGAAUUUGAAACCGAA ((((((.((...((((((.....))))))...)).)))))).((...((((((...((((((((....((....))...))))).)))..))))))..((((.......))))....)). (-37.43 = -38.10 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:09 2006