
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,937,313 – 20,937,713 |
| Length | 400 |
| Max. P | 0.974505 |

| Location | 20,937,313 – 20,937,433 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -35.53 |
| Energy contribution | -36.53 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20937313 120 + 22407834 UUUAACGUCAUGGCCAAUCCUUGUUCCCGGAGGCGGUUCGGAAAUACCGAAAUCUCGGCCUGAAACAUUGAUAAGUGGCUGGCCAGAGAAUCGAAAUUGGCCAAAGACGCAGAGCGAUGU ....(((((.((((((((..((((((.(.((((...(((((.....))))).))))((((.(...((((....)))).).)))).).))).))).)))))))).....((...))))))) ( -39.50) >DroSec_CAF1 23572 120 + 1 AUUAACGUCAUGGCCAAUCCUUGUUCCCGGAGGCUGUUCGGAAAUACCGAAAUCCCGGCCUGAAACAUUGAUAAGUGGCUGGCCGGAGAAUCGAAAUUGGCCAAAGACGCAGAGCGAUGU ....(((((.((((((((..(((((((((((((((((((((.....)))))....))))))....((((....)))).....)))).))).))).)))))))).....((...))))))) ( -42.50) >DroSim_CAF1 21932 120 + 1 AUUAACGUCAUGGCAAAUCCUUGUUCCCGGAGGCGGUUCGGAAAUACCGAAAUCUCGGCCUGAAACAUUGAUAAGUGCCUGGCCGGAGAAUCGAAAUUGGCCAAAGACGCAGAGCGAUCU .....((((..((((.(((..((((..(((.(.((((((((.....)))))...))).)))).))))..)))...))))(((((((..........)))))))..))))........... ( -34.40) >consensus AUUAACGUCAUGGCCAAUCCUUGUUCCCGGAGGCGGUUCGGAAAUACCGAAAUCUCGGCCUGAAACAUUGAUAAGUGGCUGGCCGGAGAAUCGAAAUUGGCCAAAGACGCAGAGCGAUGU .....((((.((((((((..((((((((.((((...(((((.....))))).))))((((.(...((((....)))).).)))))).))).))).))))))))..))))........... (-35.53 = -36.53 + 1.00)

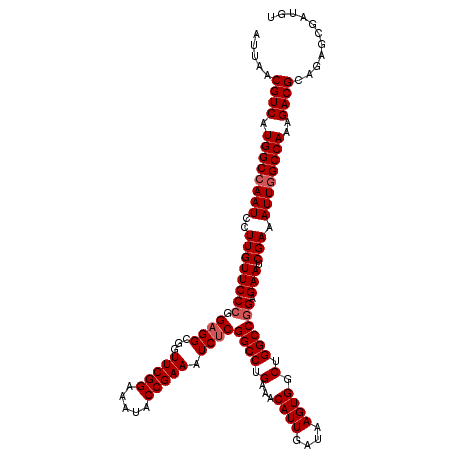
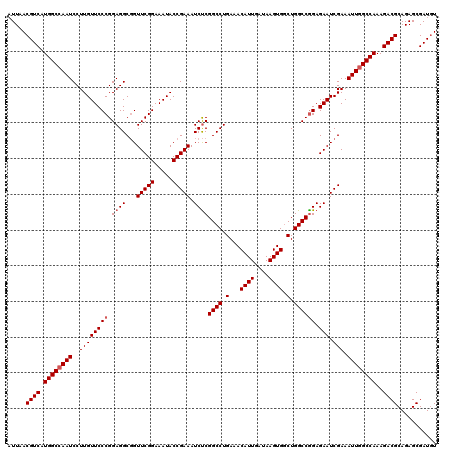
| Location | 20,937,313 – 20,937,433 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -38.29 |
| Energy contribution | -38.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20937313 120 - 22407834 ACAUCGCUCUGCGUCUUUGGCCAAUUUCGAUUCUCUGGCCAGCCACUUAUCAAUGUUUCAGGCCGAGAUUUCGGUAUUUCCGAACCGCCUCCGGGAACAAGGAUUGGCCAUGACGUUAAA ..........(((((..(((((((((((((.((((.((((.(..((........))..).))))))))..)))...((((((.........))))))...)))))))))).))))).... ( -40.00) >DroSec_CAF1 23572 120 - 1 ACAUCGCUCUGCGUCUUUGGCCAAUUUCGAUUCUCCGGCCAGCCACUUAUCAAUGUUUCAGGCCGGGAUUUCGGUAUUUCCGAACAGCCUCCGGGAACAAGGAUUGGCCAUGACGUUAAU ..........(((((..((((((((((...((((((((((.(..((........))..).)))))((..(((((.....)))))...))...)))))...)))))))))).))))).... ( -42.40) >DroSim_CAF1 21932 120 - 1 AGAUCGCUCUGCGUCUUUGGCCAAUUUCGAUUCUCCGGCCAGGCACUUAUCAAUGUUUCAGGCCGAGAUUUCGGUAUUUCCGAACCGCCUCCGGGAACAAGGAUUUGCCAUGACGUUAAU ..........(((((..((((.(((((...(((.((((..((((..........(((((.....)))))(((((.....)))))..)))))))))))...))))).)))).))))).... ( -36.60) >consensus ACAUCGCUCUGCGUCUUUGGCCAAUUUCGAUUCUCCGGCCAGCCACUUAUCAAUGUUUCAGGCCGAGAUUUCGGUAUUUCCGAACCGCCUCCGGGAACAAGGAUUGGCCAUGACGUUAAU ..........(((((..(((((((((((((.((((.((((.(..((........))..).))))))))..)))...((((((.........))))))...)))))))))).))))).... (-38.29 = -38.40 + 0.11)

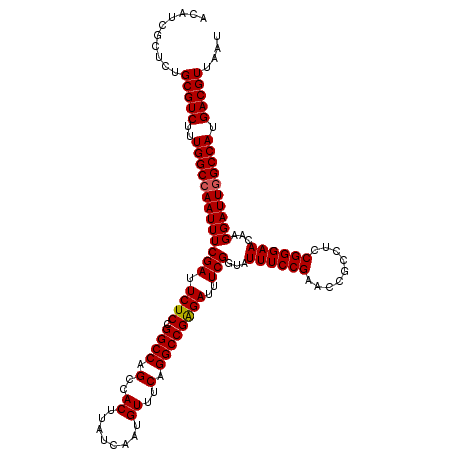

| Location | 20,937,353 – 20,937,473 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -35.74 |
| Energy contribution | -35.63 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20937353 120 + 22407834 GAAAUACCGAAAUCUCGGCCUGAAACAUUGAUAAGUGGCUGGCCAGAGAAUCGAAAUUGGCCAAAGACGCAGAGCGAUGUGCUGGCCACAGUCAUAAUUAAUACAGGAGGCCGGCAAACA ..............(((((((.....((((((..((((((((((((...((((...((.((.......)).)).))))...))))))..)))))).)))))).....)))))))...... ( -37.00) >DroSec_CAF1 23612 120 + 1 GAAAUACCGAAAUCCCGGCCUGAAACAUUGAUAAGUGGCUGGCCGGAGAAUCGAAAUUGGCCAAAGACGCAGAGCGAUGUGCUGGCCACAGUCAUAAUUAAUACAGGAGGCCGGCAAACA ..............(((((((.....((((((..((((((((((((...((((...((.((.......)).)).))))...))))))..)))))).)))))).....)))))))...... ( -38.70) >DroSim_CAF1 21972 120 + 1 GAAAUACCGAAAUCUCGGCCUGAAACAUUGAUAAGUGCCUGGCCGGAGAAUCGAAAUUGGCCAAAGACGCAGAGCGAUCUGCUGGCCACAGUCAUAAUUAAUACAGGAGGCCGGCAAACA ..............(((((((....((((....))))((((....((..((.((...((((((.....(((((....)))))))))))...))))..))....)))))))))))...... ( -35.40) >consensus GAAAUACCGAAAUCUCGGCCUGAAACAUUGAUAAGUGGCUGGCCGGAGAAUCGAAAUUGGCCAAAGACGCAGAGCGAUGUGCUGGCCACAGUCAUAAUUAAUACAGGAGGCCGGCAAACA ..............(((((((.....((((((..((((((((((((...((((...((.((.......)).)).))))...))))))..)))))).)))))).....)))))))...... (-35.74 = -35.63 + -0.11)


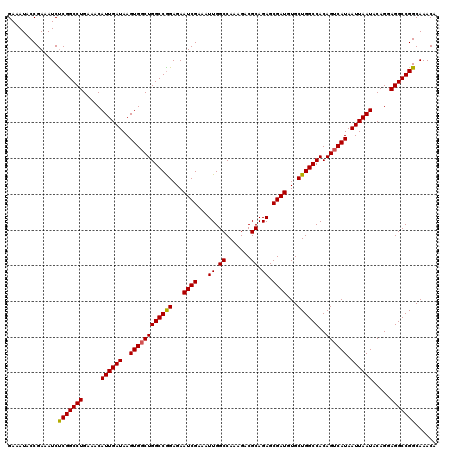
| Location | 20,937,433 – 20,937,553 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -32.70 |
| Energy contribution | -33.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20937433 120 - 22407834 GCAAGGCAAAUGCAAAUGCAAAUGCAGACGUUGCUGGCAUGUGUGGUAUAAUAUAUGUGUACUUUAUUUGUUUGCCACUUUGUUUGCCGGCCUCCUGUAUUAAUUAUGACUGUGGCCAGC ((..(((((((((((.(((....)))....))))(((((.(((.((((((.......)))))).))).....)))))....)))))))((((.(...(((.....)))...).)))).)) ( -35.30) >DroSec_CAF1 23692 119 - 1 GCAAGGCAAAUGCAAAUGCAAAUGCAGAUGUUGCUUGCAUGUGU-GUAUAAUAUAUGUGGACUUUAUUUGUUUGCCACUUUGUUUGCCGGCCUCCUGUAUUAAUUAUGACUGUGGCCAGC ((..(((((..(((((.((((..(((((((..(.(..(((((((-.....)))))))..).)..)))))))))))...))))))))))((((.(...(((.....)))...).)))).)) ( -37.20) >DroSim_CAF1 22052 119 - 1 GCAAGGCAAAUGCAAAUGCAAAUGCAGAUGUUGCUGGCAUGUGU-GUAUAAUAUAUGUGGACUUUAUUUGUUUGCCACUUUGUUUGCCGGCCUCCUGUAUUAAUUAUGACUGUGGCCAGC ((..(((((..(((((.((((..(((((((..(.(.((((((((-.....)))))))).).)..)))))))))))...))))))))))((((.(...(((.....)))...).)))).)) ( -36.20) >consensus GCAAGGCAAAUGCAAAUGCAAAUGCAGAUGUUGCUGGCAUGUGU_GUAUAAUAUAUGUGGACUUUAUUUGUUUGCCACUUUGUUUGCCGGCCUCCUGUAUUAAUUAUGACUGUGGCCAGC ((..(((((..(((((.((((..(((((((..(.(.((((((((......)))))))).).)..)))))))))))...))))))))))((((.(...(((.....)))...).)))).)) (-32.70 = -33.37 + 0.67)

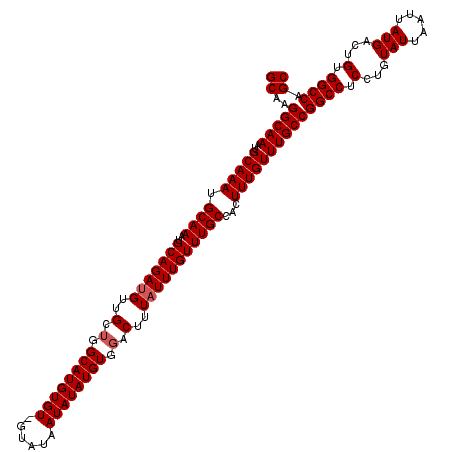
| Location | 20,937,473 – 20,937,593 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -19.47 |
| Energy contribution | -19.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20937473 120 + 22407834 AAGUGGCAAACAAAUAAAGUACACAUAUAUUAUACCACACAUGCCAGCAACGUCUGCAUUUGCAUUUGCAUUUGCCUUGCAGUGUGUUGUCCUAACGCCCCCUCUCAAAAUUUAAACAUA .((.(((((....((((.(((....))).))))..(((((.(((.((........(((..(((....)))..))))).)))))))))))))))........................... ( -20.50) >DroSec_CAF1 23732 119 + 1 AAGUGGCAAACAAAUAAAGUCCACAUAUAUUAUAC-ACACAUGCAAGCAACAUCUGCAUUUGCAUUUGCAUUUGCCUUGCAGUGUGUUGUCCCAACGCCCCCUCUCAAAAUUUAAACACA ..(((((...........).)))).........((-((((.((((((........(((..(((....)))..)))))))))))))))................................. ( -25.40) >DroSim_CAF1 22092 119 + 1 AAGUGGCAAACAAAUAAAGUCCACAUAUAUUAUAC-ACACAUGCCAGCAACAUCUGCAUUUGCAUUUGCAUUUGCCUUGCAGUGUGUUGUCCCAACGCCCCCUCUCAAAAUUUAAACACA ..(((((...........).)))).........((-((((.(((.((........(((..(((....)))..))))).)))))))))................................. ( -21.30) >consensus AAGUGGCAAACAAAUAAAGUCCACAUAUAUUAUAC_ACACAUGCCAGCAACAUCUGCAUUUGCAUUUGCAUUUGCCUUGCAGUGUGUUGUCCCAACGCCCCCUCUCAAAAUUUAAACACA ....(((......((((.((......)).))))((.((((((....((((.....(((..(((....)))..))).)))).)))))).))......)))..................... (-19.47 = -19.47 + -0.00)



| Location | 20,937,593 – 20,937,713 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -29.59 |
| Energy contribution | -29.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.47 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20937593 120 + 22407834 CACUUUAGUUAUUUAUUGCUGCUCUCGUUUGCCCAUCGAUGUUGUCUUGUUGUGCGGCAAAUGCAUUUUAAUAUUUCACAUUGAUUUGUUAUGCAAAAAUUUGAUGGCUCUGUGAUACAC (((...(((((((.(((..(((...((((((((((.(((((......))))))).)))))))).....(((((..((.....))..))))).)))..)))..)))))))..)))...... ( -27.70) >DroSec_CAF1 23851 120 + 1 CACUUUAGUUAUUUAUUGCUGCUCUCGUUUGCCCAUCGACGUUGUCUUGUUGUGCGGCAAAUGCAUUUUAAUAUUUCACAUUGAUUUGUUAUGCAAAAAUUUGAUGGCUCUGUGAUACAC (((...(((((((.(((..(((...((((((((((.(((((......))))))).)))))))).....(((((..((.....))..))))).)))..)))..)))))))..)))...... ( -30.20) >DroSim_CAF1 22211 120 + 1 CACUUUAGUUAUUUAUUGCUGCUCUCGUUUGCCCAUCGACGUUGUCUUGUUGUGCGGCAAAUGCAUUUUAAUAUUUCACAUUGAUUUGUUAUGCAAAAAUUUGAUGGCUCUGUGAUACAC (((...(((((((.(((..(((...((((((((((.(((((......))))))).)))))))).....(((((..((.....))..))))).)))..)))..)))))))..)))...... ( -30.20) >consensus CACUUUAGUUAUUUAUUGCUGCUCUCGUUUGCCCAUCGACGUUGUCUUGUUGUGCGGCAAAUGCAUUUUAAUAUUUCACAUUGAUUUGUUAUGCAAAAAUUUGAUGGCUCUGUGAUACAC (((...(((((((.(((..(((...((((((((((.(((((......))))))).)))))))).....(((((..((.....))..))))).)))..)))..)))))))..)))...... (-29.59 = -29.37 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:58 2006