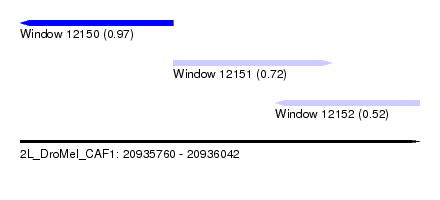
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,935,760 – 20,936,042 |
| Length | 282 |
| Max. P | 0.971001 |

| Location | 20,935,760 – 20,935,868 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 98.77 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -22.73 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20935760 108 - 22407834 CUGCUCCAAAUUCGAAUGUGAAACAUGGAAAAAGUUAUACCAAACAUGGAAAUGUAUGUCAUUGACUAAAUGACAAAUCACUUGACAGUUUUUCUAUUCUCACACAAU ................(((((...(((((((((((((......((((....)))).(((((((.....))))))).......))))..)))))))))..))))).... ( -22.90) >DroSec_CAF1 22105 108 - 1 CUGCUCCAAAUUCGAAUGUGCAACAUGGAAAAAGUUAUACCAAACAUGGAAAUGUAUGUCAUUGACUAAAUGACAAAUCACUUGACAGUUUUUCUAUUCUCGCACAAU ................(((((...(((((((((((((......((((....)))).(((((((.....))))))).......))))..)))))))))....))))).. ( -24.50) >DroSim_CAF1 20422 108 - 1 CUGCUCCAAAUUCGAAUGUGCAACAUGGAAAAAGUUAUACCAAACAUGGAAAUGUAUGUCAUUGACUAAAUGACAAAUCACUUGACAGUUUUUCUAUUCUCGCACAAU ................(((((...(((((((((((((......((((....)))).(((((((.....))))))).......))))..)))))))))....))))).. ( -24.50) >consensus CUGCUCCAAAUUCGAAUGUGCAACAUGGAAAAAGUUAUACCAAACAUGGAAAUGUAUGUCAUUGACUAAAUGACAAAUCACUUGACAGUUUUUCUAUUCUCGCACAAU ................(((((...(((((((((((((......((((....)))).(((((((.....))))))).......))))..)))))))))....))))).. (-22.73 = -23.07 + 0.33)

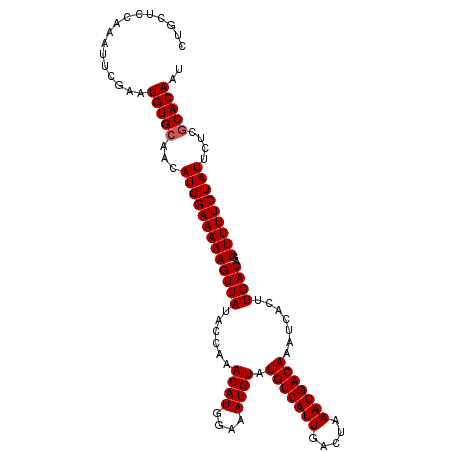
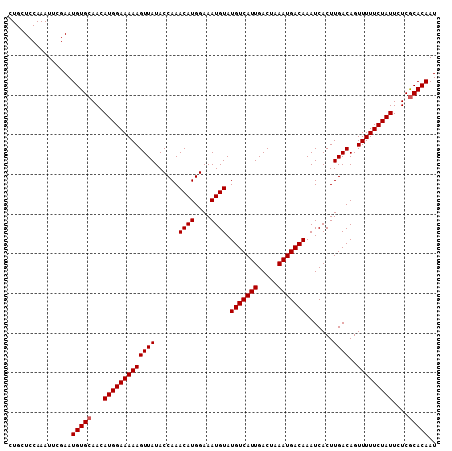
| Location | 20,935,868 – 20,935,980 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -41.11 |
| Consensus MFE | -39.04 |
| Energy contribution | -38.60 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20935868 112 + 22407834 UUCCCGGCGCUGCUGUUGUAAUUGAAGCAUGUAAUGCAGCAAACCCAACUGCAGA--------CCGGUGACCUCCGCCCCCGGCAGUUGGGGGAAUUGUCAUACAAAAGCUUUUGGGAGC (((((((....(((.(((((.(((..((((...))))..))).(((((((((...--------..((((.....))))....)))))))))..........))))).)))..))))))). ( -40.90) >DroSec_CAF1 22213 120 + 1 UUCCCGCCGCUGCUGUUGUAAUUGAAGCAUGUAAUGCAGCAAACCCAACUGCAGACCUCCCAACCGGUGACCUCCGCCCCCGGCAGUCGGGGGAAUUGUCAUACAAAAGCUUUUGGGAGC .........((((.((((...(((..((((...))))..)))...)))).))))..(((((((...(((((.....((((((.....))))))....)))))..........))))))). ( -40.22) >DroSim_CAF1 20530 120 + 1 UUCCCGCCGCUGCUGUUGUAAUUGAAGCAUGUAAUGCAGCAAACCCAACUGCAGACCUCCCAACCGGUGACCUCCGCCCCCGGCAGCCGGGGGAAUUGUCAUACAAAAGCUUUUGGGAGC .........((((.((((...(((..((((...))))..)))...)))).))))..(((((((...(((((.....(((((((...)))))))....)))))..........))))))). ( -42.22) >consensus UUCCCGCCGCUGCUGUUGUAAUUGAAGCAUGUAAUGCAGCAAACCCAACUGCAGACCUCCCAACCGGUGACCUCCGCCCCCGGCAGUCGGGGGAAUUGUCAUACAAAAGCUUUUGGGAGC ((((((...((((.((((...(((..((((...))))..)))...)))).))))............(((((.....(((((((...)))))))....)))))...........)))))). (-39.04 = -38.60 + -0.44)

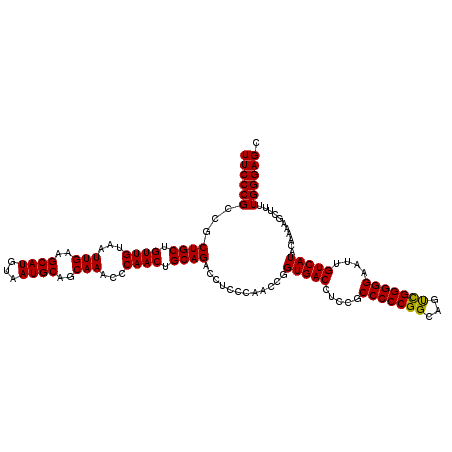

| Location | 20,935,940 – 20,936,042 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 98.69 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -30.89 |
| Energy contribution | -30.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 1.00 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20935940 102 - 22407834 GGCUUGUGGGGCGGAUUUGUUGCCUUGAGGUCAUUAUAUCGAAAACAGCAGUUGCCCAAAAUGCUCCCAAAAGCUUUUGUAUGACAAUUCCCCCAACUGCCG (((...(((((.((((((((((..((((..........))))...)))))((((..(((((.(((......))))))))..))))))))))))))...))). ( -31.50) >DroSec_CAF1 22293 102 - 1 GGCUUGUGGGGCGGAUUUGUUGCCUUGAGGUCAUUAUAUCGAAAACAGCAGUUGCCCAAAAUGCUCCCAAAAGCUUUUGUAUGACAAUUCCCCCGACUGCCG (((...(((((.((((((((((..((((..........))))...)))))((((..(((((.(((......))))))))..))))))))))))))...))). ( -30.80) >DroSim_CAF1 20610 102 - 1 GGCUUGUGGGGCGGAUUUGUUGCCUUGAGGUCAUUAUAUCGAAAACAGCAGUUGCCCAAAAUGCUCCCAAAAGCUUUUGUAUGACAAUUCCCCCGGCUGCCG (((..((.(((.(((..(((((..((((..........))))...)))))((((..(((((.(((......))))))))..))))...)))))).)).))). ( -30.60) >consensus GGCUUGUGGGGCGGAUUUGUUGCCUUGAGGUCAUUAUAUCGAAAACAGCAGUUGCCCAAAAUGCUCCCAAAAGCUUUUGUAUGACAAUUCCCCCGACUGCCG (((...(((((.((((((((((..((((..........))))...)))))((((..(((((.(((......))))))))..))))))))))))))...))). (-30.89 = -30.67 + -0.22)

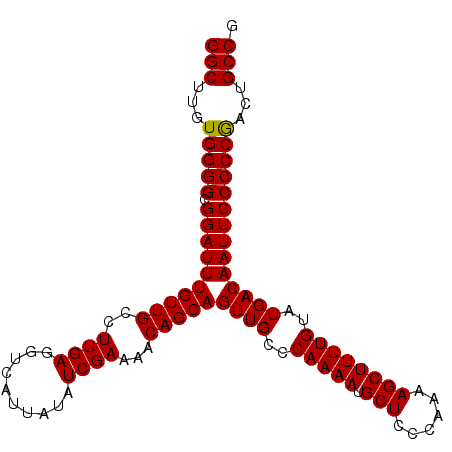
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:50 2006