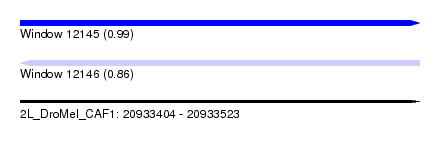
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,933,404 – 20,933,523 |
| Length | 119 |
| Max. P | 0.993793 |

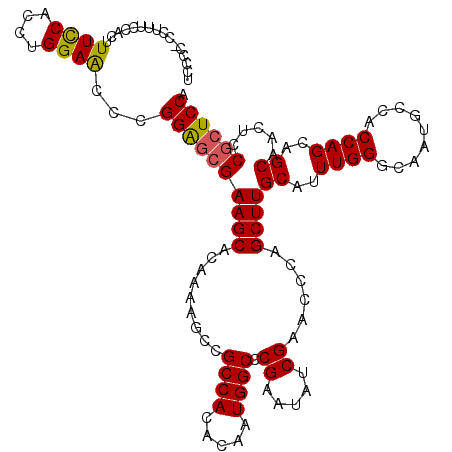
| Location | 20,933,404 – 20,933,523 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -49.00 |
| Consensus MFE | -41.78 |
| Energy contribution | -41.46 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
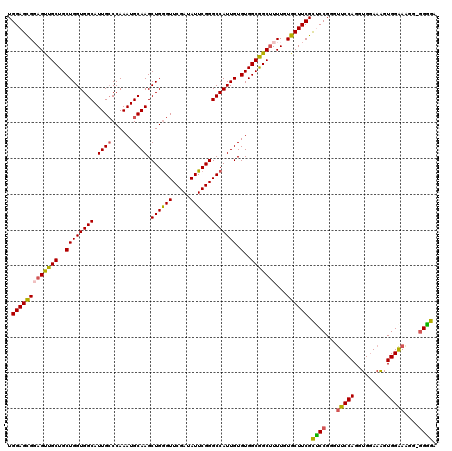
>2L_DroMel_CAF1 20933404 119 + 22407834 UGGAGCGGAGUUGCUGCUGGUGGCAUUGCCCAAAUGCAAGCUGGGUUCGAUAUUCGGGCCAUUGUGUGGCGGCUUUUGUGCUUCGUUCCGGGUUCCAGGUGAAAAGUGGAAAGG-AGGGG .((((((.((..((((((...(((.((((......))))))).((((((.....)))))).......))))))..)).)))))).((((...(((((.........))))).))-))... ( -43.00) >DroSec_CAF1 19723 119 + 1 UGGAGUGGAGUUGCUGCUGGUGGCAUUGCCCAAAUGCAAGCUGGGUUCGAUAUUCGGGCCAUUGUGUGGCAGCUUUUGUGCUUCGCCCCGGGUUCCAGGUGGAAAGUGGAAUGG-GGGGA .(((((((((((((..((((((((.((((......)))).((((((.....))))))))))))).)..))))))))...))))).((((..((((((.........))))))..-)))). ( -47.60) >DroSim_CAF1 18018 120 + 1 UGGAGCGGAGUUGCUGCUGGUGGCAUUGCCCAAAUGCAAGCUGGGUUCGAUAUUCGGGCCAUUGUGUGGCGGCUUUUGUGCUUCGCCCCGGGUUCCAGGUGGAAAGUGGAAAGGGGGGGA .((((((.((..((((((...(((.((((......))))))).((((((.....)))))).......))))))..)).)))))).((((...(((((.........))))).)))).... ( -47.60) >DroEre_CAF1 17147 118 + 1 UGGAGCGCAGCUGCUGCUGGUGGCAUUGGCCAAAUGCAAGCUGGGUUCGAUAUUCGGGCCAUUGUGUGGCGGCCUUUGUGCUUCGCUCCGGGCUCCAGGUGGAAAGUGGGAGC--GGAGA .(((((((((..((((((..((((....)))).((((((....((((((.....)))))).))))))))))))..))))))))).((((..(((((.(.(....).).)))))--)))). ( -54.20) >DroYak_CAF1 18004 118 + 1 UGGAGCGCAGCUGCUGCUGGUGGCAUUGCCCAAAUGCAAGCUGAGUUCGAUAUUCGGGCCAUUGUGUGGCGGCUUUUGUGCUUCGCUCCGGGUUCCAGGUGGAAGGUGGAAAC--GGAGA .(((((((((((((..((((((((.((((......)))).((((((.....))))))))))))).)..)))))...)))))))).(((((..(((((.........))))).)--)))). ( -52.60) >consensus UGGAGCGGAGUUGCUGCUGGUGGCAUUGCCCAAAUGCAAGCUGGGUUCGAUAUUCGGGCCAUUGUGUGGCGGCUUUUGUGCUUCGCUCCGGGUUCCAGGUGGAAAGUGGAAAGG_GGGGA .(((((((((((((..((((((((.((((......)))).((((((.....))))))))))))).)..)))))))...)))))).((((...(((((.........)))))....)))). (-41.78 = -41.46 + -0.32)

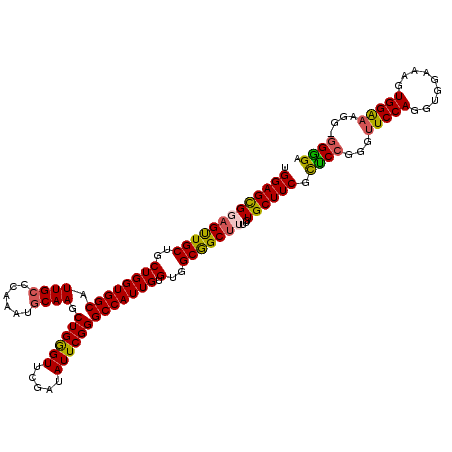
| Location | 20,933,404 – 20,933,523 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.56 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20933404 119 - 22407834 CCCCU-CCUUUCCACUUUUCACCUGGAACCCGGAACGAAGCACAAAAGCCGCCACACAAUGGCCCGAAUAUCGAACCCAGCUUGCAUUUGGGCAAUGCCACCAGCAGCAACUCCGCUCCA .....-..................(((...((((.(((.((......)).((((.....)))).......)))......(((.((.....(((...)))....)))))...)))).))). ( -23.50) >DroSec_CAF1 19723 119 - 1 UCCCC-CCAUUCCACUUUCCACCUGGAACCCGGGGCGAAGCACAAAAGCUGCCACACAAUGGCCCGAAUAUCGAACCCAGCUUGCAUUUGGGCAAUGCCACCAGCAGCAACUCCACUCCA (((((-(..(((((.........)))))...)))).)).........(((((.........((((((((..(((.......))).))))))))..........)))))............ ( -30.91) >DroSim_CAF1 18018 120 - 1 UCCCCCCCUUUCCACUUUCCACCUGGAACCCGGGGCGAAGCACAAAAGCCGCCACACAAUGGCCCGAAUAUCGAACCCAGCUUGCAUUUGGGCAAUGCCACCAGCAGCAACUCCGCUCCA ................((((....))))...((((((..((......)).((.........((((((((..(((.......))).))))))))..(((.....))))).....)))))). ( -29.00) >DroEre_CAF1 17147 118 - 1 UCUCC--GCUCCCACUUUCCACCUGGAGCCCGGAGCGAAGCACAAAGGCCGCCACACAAUGGCCCGAAUAUCGAACCCAGCUUGCAUUUGGCCAAUGCCACCAGCAGCAGCUGCGCUCCA .....--(((((............)))))..((((((.(((.....(((((........)))))...............(((.((...((((....))))...))))).))).)))))). ( -41.00) >DroYak_CAF1 18004 118 - 1 UCUCC--GUUUCCACCUUCCACCUGGAACCCGGAGCGAAGCACAAAAGCCGCCACACAAUGGCCCGAAUAUCGAACUCAGCUUGCAUUUGGGCAAUGCCACCAGCAGCAGCUGCGCUCCA .....--.........((((....))))...((((((.(((....((((.((((.....)))).((.....))......))))((..((((.........))))..)).))).)))))). ( -33.00) >consensus UCCCC_CCUUUCCACUUUCCACCUGGAACCCGGAGCGAAGCACAAAAGCCGCCACACAAUGGCCCGAAUAUCGAACCCAGCUUGCAUUUGGGCAAUGCCACCAGCAGCAACUCCGCUCCA ................((((....))))...((((((((((.........((((.....)))).((.....))......))))((..((((.........))))..)).....)))))). (-25.72 = -25.56 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:44 2006