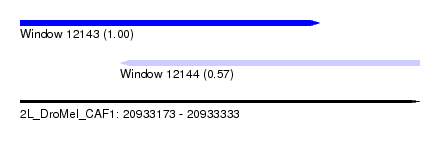
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,933,173 – 20,933,333 |
| Length | 160 |
| Max. P | 0.995499 |

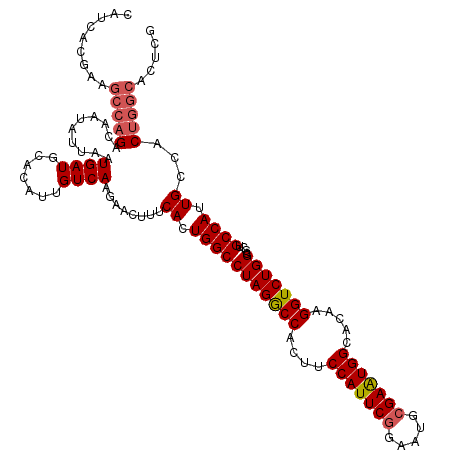
| Location | 20,933,173 – 20,933,293 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -48.53 |
| Consensus MFE | -42.72 |
| Energy contribution | -42.85 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20933173 120 + 22407834 GGGAUGGCGCGGGGUCACAGCUAGACUCAGAGGCAUGGAGCGAGUGCCAGUGGCAAUGGCAGCGCCAGACCUUGUGCCAUUCGCAUUCCGAAUGGAAGUGGCCUAGGCCAGUGAAAGUUC .(((((((((((((((...(((.........))).(((.((...(((((.......))))))).))))))))))))))))))((.((((....)))).((((....))))))........ ( -50.50) >DroSim_CAF1 17787 120 + 1 GGUAUGGCGCGGGGUCACAGCUAGAGUCAGAGGCAUAGAGCGAGUGCCAGUGGCAAUGGCAGCGCCAGACCUUGUGCCAUUCGCAUUCCGAAUGGAAGUGGCCUAGGCCAGUGAAAGUUC ....((((((((((((.........((((..(((((.......)))))..))))..((((...))))))))))))))))(((((.((((....)))).((((....)))))))))..... ( -51.90) >DroEre_CAF1 16949 90 + 1 GGAAUGGUGCAGGGUCACA------------------------------GUGGCAAUGGCUGCGCCAGACCUUGUGCCACUCGCAUUCCGAAUGGAAGUGGUCUAGGCCAGUGAAAGUUC ....(((..(((((((...------------------------------(..((....))..)....)))))))..))).((((.((((....)))).((((....))))))))...... ( -36.50) >DroYak_CAF1 17768 120 + 1 GGAAUGGCGCGGGGUCACAGCUCGAGUCAGAGGCAUAGAGCGAGUGCCAGUGGCAAUGGCAGCGCCAGACCUUGUGCCAUUCCCAUUCCGAAUGGAAGUGGCCUAGGCCAGUGAAAGUUC ((((((((((((((((.........((((..(((((.......)))))..))))..((((...))))))))))))))))))))......((((...(.((((....)))).)....)))) ( -55.20) >consensus GGAAUGGCGCGGGGUCACAGCUAGAGUCAGAGGCAUAGAGCGAGUGCCAGUGGCAAUGGCAGCGCCAGACCUUGUGCCAUUCGCAUUCCGAAUGGAAGUGGCCUAGGCCAGUGAAAGUUC .(((((((((((((((...............(((((.......)))))..((((.........))))))))))))))))))).......((((...(.((((....)))).)....)))) (-42.72 = -42.85 + 0.13)

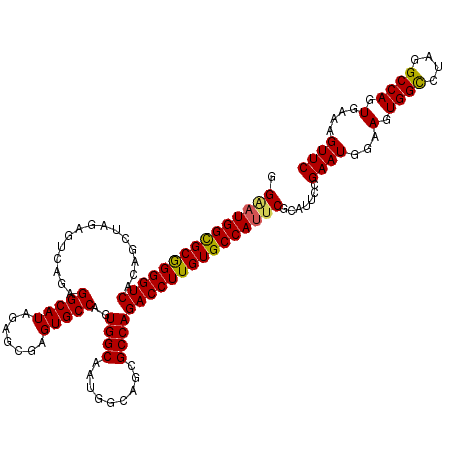
| Location | 20,933,213 – 20,933,333 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -29.80 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20933213 120 - 22407834 CAUCACGAAGCCAGACAAUAUUAAUGAUGCACAUUGUCAAGAACUUUCACUGGCCUAGGCCACUUCCAUUCGGAAUGCGAAUGGCACAAGGUCUGGCGCUGCCAUUGCCACUGGCACUCG .....(((.(((((((........((((.......))))...........((((....))))((((((((((.....)))))))...))))))))))..(((((.......))))).))) ( -37.20) >DroSim_CAF1 17827 120 - 1 CAUCACGAAGCCAGACAAUAUUAAUGAUGCACAUUGUCAAGAACUUUCACUGGCCUAGGCCACUUCCAUUCGGAAUGCGAAUGGCACAAGGUCUGGCGCUGCCAUUGCCACUGGCACUCG .....(((.(((((((........((((.......))))...........((((....))))((((((((((.....)))))))...))))))))))..(((((.......))))).))) ( -37.20) >DroEre_CAF1 16968 111 - 1 CAUCACGAAGCCAGACAAUAUUAAUGAUGCACAUUGUCAAGAACUUUCACUGGCCUAGACCACUUCCAUUCGGAAUGCGAGUGGCACAAGGUCUGGCGCAGCCAUUGCCAC--------- .........((..((((((.............))))))...........((((((.(((((....(((((((.....))))))).....)))))))).))).....))...--------- ( -29.42) >DroYak_CAF1 17808 120 - 1 CAUCACGAAGCCAGACAAUAUUAAUGAUGCACAUUGUCAAGAACUUUCACUGGCCUAGGCCACUUCCAUUCGGAAUGGGAAUGGCACAAGGUCUGGCGCUGCCAUUGCCACUGGCACUCG .....(((.(((((((........((.(((.((((.(((...........((((....)))).((((....))))))).)))))))))..)))))))..(((((.......))))).))) ( -37.30) >consensus CAUCACGAAGCCAGACAAUAUUAAUGAUGCACAUUGUCAAGAACUUUCACUGGCCUAGGCCACUUCCAUUCGGAAUGCGAAUGGCACAAGGUCUGGCGCUGCCAUUGCCACUGGCACUCG .........(((((..........((((.......))))........((.(((((((((((....(((((((.....))))))).....)))))))....)))).))...)))))..... (-29.80 = -30.67 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:42 2006