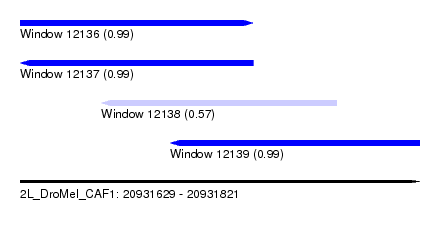
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,931,629 – 20,931,821 |
| Length | 192 |
| Max. P | 0.992512 |

| Location | 20,931,629 – 20,931,741 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -21.55 |
| Energy contribution | -22.67 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20931629 112 + 22407834 CCACCGCUCACAUUUUCCUCAUUUUCCCCAUCGCCUGAUGGCGGAGGCGGCUUUCCACC-------GGAUUCUUUUCGCUUUUCUGCGCCGCGAACGCUGCUGCUGAUUUUCACUUCUU .....((.................((((((((....))))).)))((((((.(((...(-------((........(((......)))))).))).))))))))............... ( -27.40) >DroSec_CAF1 18050 110 + 1 CCACCGCUCACAUUUUCCUCAUUUUCCCCAUCGCCUGAUGGUGGAGGCAGCU--CCACC-------GGCUUCUUUUCGCUUUUAUGCGCCGCCACCGCUUCUGCUGAUUUUCACUUCUU .............................(((((..((((((((.(((((((--.....-------)))).......((......))))).))))))..)).)).)))........... ( -23.20) >DroEre_CAF1 15556 108 + 1 -----------AUUUUCCUUAUUUUCCCCAUCGCUUGAUGGCGGAGGCGGCUUUCCACCGAAAGGCGGAUUUCUUCCGCUUUUCCACGCCGCCAGCGCUGCUGCUGAUUUUCACUUCUU -----------.............((((((((....))))).)))((((((........((((((((((.....))))))))))...))))))((((....)))).............. ( -38.60) >consensus CCACCGCUCACAUUUUCCUCAUUUUCCCCAUCGCCUGAUGGCGGAGGCGGCUUUCCACC_______GGAUUCUUUUCGCUUUUCUGCGCCGCCAACGCUGCUGCUGAUUUUCACUUCUU ........................((((((((....))))).)))((((((...............(((.....)))((......))))))))...((....))............... (-21.55 = -22.67 + 1.11)

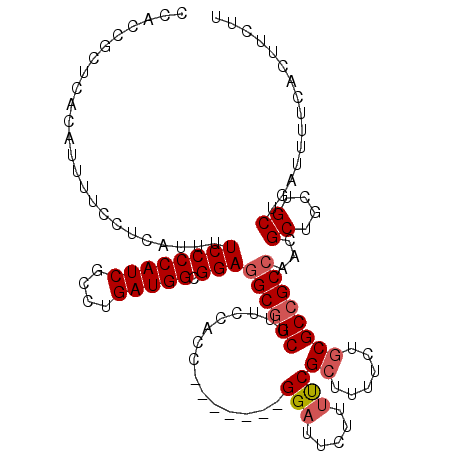
| Location | 20,931,629 – 20,931,741 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -23.81 |
| Energy contribution | -23.87 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20931629 112 - 22407834 AAGAAGUGAAAAUCAGCAGCAGCGUUCGCGGCGCAGAAAAGCGAAAAGAAUCC-------GGUGGAAAGCCGCCUCCGCCAUCAGGCGAUGGGGAAAAUGAGGAAAAUGUGAGCGGUGG ...................((.((((((((.(((......))).......(((-------(((((....)))))(((.(((((....))))))))......)))...)))))))).)). ( -38.70) >DroSec_CAF1 18050 110 - 1 AAGAAGUGAAAAUCAGCAGAAGCGGUGGCGGCGCAUAAAAGCGAAAAGAAGCC-------GGUGG--AGCUGCCUCCACCAUCAGGCGAUGGGGAAAAUGAGGAAAAUGUGAGCGGUGG ............(((((((.(.((((..(..(((......)))....)..)))-------).)..--..)))).(((.(((((....))))))))...))).................. ( -28.90) >DroEre_CAF1 15556 108 - 1 AAGAAGUGAAAAUCAGCAGCAGCGCUGGCGGCGUGGAAAAGCGGAAGAAAUCCGCCUUUCGGUGGAAAGCCGCCUCCGCCAUCAAGCGAUGGGGAAAAUAAGGAAAAU----------- ..............(((......)))((((((.........(....)...((((((....))))))..))))))(((.(((((....)))))))).............----------- ( -37.00) >consensus AAGAAGUGAAAAUCAGCAGCAGCGCUGGCGGCGCAGAAAAGCGAAAAGAAUCC_______GGUGGAAAGCCGCCUCCGCCAUCAGGCGAUGGGGAAAAUGAGGAAAAUGUGAGCGGUGG ...............((....))...((((((((......)).........(((.....)))......))))))(((.(((((....))))))))........................ (-23.81 = -23.87 + 0.06)

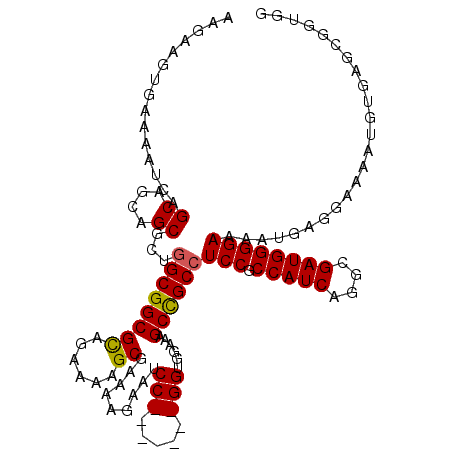
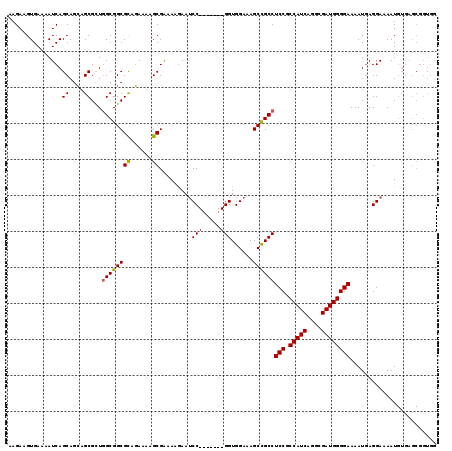
| Location | 20,931,668 – 20,931,781 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -25.46 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20931668 113 - 22407834 UUGUUGCGGUUUUGACUUAUGCGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGUUCGCGGCGCAGAAAAGCGAAAAGAAUCC-------GGUGGAAAGCCGCCUCCGCC .....(((((((((.(((.(((((..(((((((....))))))).(((((.....((....)).))))).)))))...)))))))).......-------(((((....))))).)))). ( -37.20) >DroSec_CAF1 18089 111 - 1 UUGUUGCGGUUUUGACUUAUGCGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGAAGCGGUGGCGGCGCAUAAAAGCGAAAAGAAGCC-------GGUGG--AGCUGCCUCCACC ..((..(.(((((..((.((...((((((((((....)))))).))))...)).))..))))).)..))(((..................)))-------(((((--((....))))))) ( -35.97) >DroSim_CAF1 16301 113 - 1 UUGUUGCGGUUUUGACUUAUGCGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGUUGGCGGCGCAGAAAAGCGAAAAGAAGCC-------GGUGGAAAGCCGCCUCCACC (((((((((((((.(((.........(((((((....)))))))))).)))))).)))))))....(((..(((......))).......)))-------((((((........)))))) ( -34.70) >DroEre_CAF1 15584 120 - 1 UUGUUGCGGUUUUGACUUAUGUGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGCUGGCGGCGUGGAAAAGCGGAAGAAAUCCGCCUUUCGGUGGAAAGCCGCCUCCGCC .((((((((((((.(((.........(((((((....)))))))))).)))))).))))))(((..((((((.........(....)...((((((....))))))..))))))..))). ( -48.30) >DroYak_CAF1 16286 93 - 1 UUGUUGCGGUUUUGACUUAUGUGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGUUGGCGUCGCACAAAAACGCAAAGAAUAC--------------------------- .((((((((((((.(((.........(((((((....)))))))))).)))))).))))))(((((.((...)).....))))).........--------------------------- ( -24.50) >consensus UUGUUGCGGUUUUGACUUAUGCGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGUUGGCGGCGCAGAAAAGCGAAAAGAAUCC_______GGUGGAAAGCCGCCUCCACC .((((((((((((.(((.........(((((((....)))))))))).)))))).))))))((((.....))))..........................((((((........)))))) (-25.46 = -26.16 + 0.70)

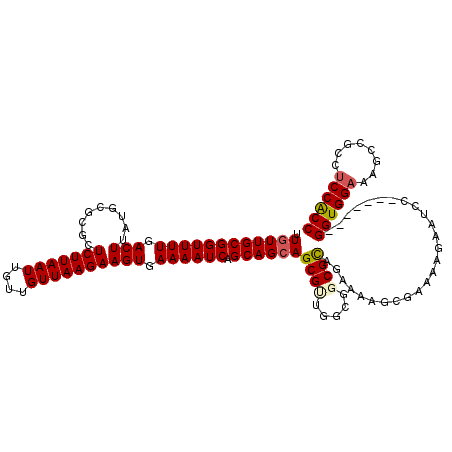
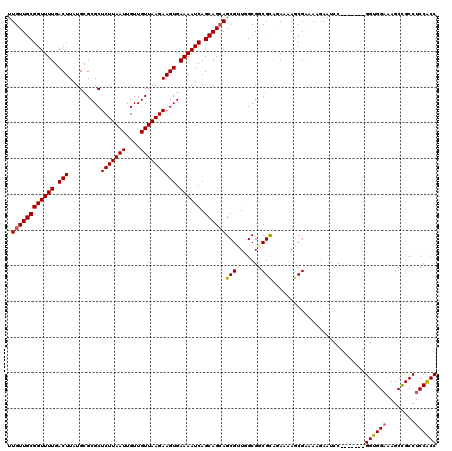
| Location | 20,931,701 – 20,931,821 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -33.40 |
| Energy contribution | -33.16 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20931701 120 - 22407834 UUGUUGCUGUAUUCACAUUGGUUGCUCCAUUUCGCCGUUGUUGUUGCGGUUUUGACUUAUGCGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGUUCGCGGCGCAGAAAA ......((((........(((.....)))....((((((((((((((((((((.(((.........(((((((....)))))))))).)))))).)))))))))...))))))))).... ( -37.40) >DroSec_CAF1 18120 120 - 1 UUGUUGCUGUAUUCACAUUGGUUGCUCCAUUUCGCCGGUGUUGUUGCGGUUUUGACUUAUGCGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGAAGCGGUGGCGGCGCAUAAAA ..(((((((((...((((((((...........))))))))...))))))...)))(((((((((((((((((....))))))).......(((.((....))))).)).)))))))).. ( -33.70) >DroSim_CAF1 16334 120 - 1 UUGUUGCCGUAUUCACAUUGGUUGCUCCAUUUCGCCGUUGUUGUUGCGGUUUUGACUUAUGCGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGUUGGCGGCGCAGAAAA ((((.(((..........(((.....)))....((((.(((((((((((((((.(((.........(((((((....)))))))))).)))))).))))))))).))))))))))).... ( -39.70) >DroEre_CAF1 15624 120 - 1 UUGUUGCUGUGUUCACAUUGGUUGCUCCAUUUCGCUGUUGUUGUUGCGGUUUUGACUUAUGUGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGCUGGCGGCGUGGAAAA ..((.((((((....)).)))).))(((((.(((((..(((((((((((((((.(((.........(((((((....)))))))))).)))))).)))))))))..))))).)))))... ( -40.20) >DroYak_CAF1 16299 120 - 1 UUGUUGCUGCGUUCACAUUGGUUUCUCUAUUUCGCUGUUGUUGUUGCGGUUUUGACUUAUGUGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGUUGGCGUCGCACAAAA ...(((.((((.......(((.....)))...((((..(((((((((((((((.(((.........(((((((....)))))))))).)))))).)))))))))..)))).))))))).. ( -34.40) >consensus UUGUUGCUGUAUUCACAUUGGUUGCUCCAUUUCGCCGUUGUUGUUGCGGUUUUGACUUAUGCGCGCUCUUAAUUGUUGUUAAGAAGUGAAAAUCAGCAGCAGCGUUGGCGGCGCAGAAAA ....(((...........(((.....)))...(((((.(((((((((((((((.(((.........(((((((....)))))))))).)))))).))))))))).)))))..)))..... (-33.40 = -33.16 + -0.24)

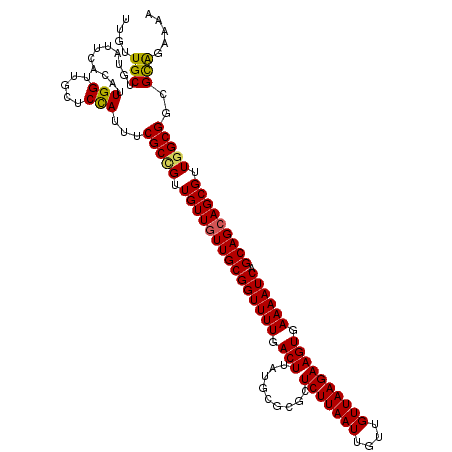
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:38 2006