
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,918,596 – 20,918,756 |
| Length | 160 |
| Max. P | 0.999814 |

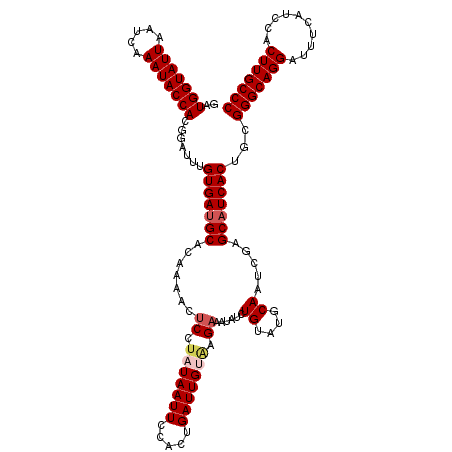
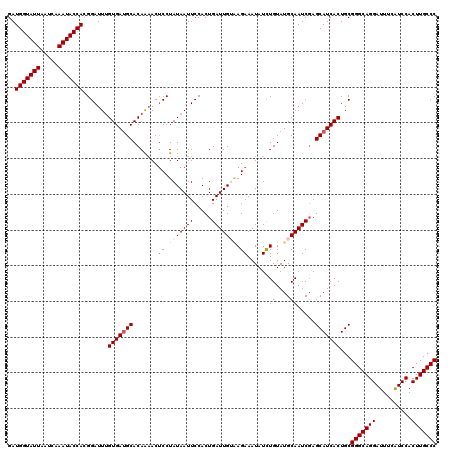
| Location | 20,918,596 – 20,918,716 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -26.58 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20918596 120 + 22407834 GAUGGUAUUUAUCAAAUACCACGGAUUUGUGAUGCACAGAACUCCUAUAAUUCCACUGAUUGUAAGAAAUAUCUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCC ..(((((((.....))))))).......(((((((..((.....))..........(((((((((((....)))...)))))))).)))))))...(((((((..........))))))) ( -32.70) >DroSec_CAF1 3950 120 + 1 GAUGGUAUUAAUCAAAUACCACGGAUUUGUGACGCACAAAACUCCUAUAAUUCCACUGAUUGAAAGAAAUAUUUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCC ..(((((((.....))))))).(((((((((...))))))..)))...........((((((..(((....))).....)))))).(((....)))(((((((..........))))))) ( -26.50) >DroSim_CAF1 3210 120 + 1 GAUGGUAUUAAUCAAAUACCACGGAUUUGUGAUGCACAGAACUCCUAUAAUUCCACUGAUUGACAGAAAUAUUUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCC ..(((((((.....))))))).......(((((((..((.....))..........(((((((((((....)))))...)))))).)))))))...(((((((..........))))))) ( -31.60) >DroEre_CAF1 3004 120 + 1 GAUGGUAUUAAUCAAAUACCAGGGAUUGGUGAUGCACAAAACCCCUAUAAUUCCACUGAUUGUGAGAAAUAUCUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCGUCCACUUGCCC ..(((((((.....)))))))......((((((((...............((((((.....))).))).....((....)).....))))))))..(((((((..........))))))) ( -32.40) >DroYak_CAF1 3007 120 + 1 GAUGGUAUUAAUCAAAUACCAGGGAUUUGUGAUGCACAAUACUCCUAUAAUUCCACUGAUUGUGAGAAAUAUCUGUAUGCAAUCUAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCC ..(((((((.....))))))).......(((((((...((((....(((.((((((.....))).))).)))..))))........)))))))...(((((((..........))))))) ( -31.70) >consensus GAUGGUAUUAAUCAAAUACCACGGAUUUGUGAUGCACAAAACUCCUAUAAUUCCACUGAUUGUAAGAAAUAUCUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCC ..(((((((.....))))))).......(((((((.......((.(((((((.....))))))).))......((....)).....)))))))...(((((((..........))))))) (-26.58 = -27.42 + 0.84)

| Location | 20,918,596 – 20,918,716 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -33.31 |
| Energy contribution | -32.95 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20918596 120 - 22407834 GGGCAAGUGGAUGAAAUCCUGCCCGCAGUGAUGCUCGAUUGCAUACAGAUAUUUCUUACAAUCAGUGGAAUUAUAGGAGUUCUGUGCAUCACAAAUCCGUGGUAUUUGAUAAAUACCAUC (((((...((((...)))))))))...(((((((..(((((.....(((....)))..))))).(..(((((.....)))))..))))))))......((((((((.....)))))))). ( -37.40) >DroSec_CAF1 3950 120 - 1 GGGCAAGUGGAUGAAAUCCUGCCCGCAGUGAUGCUCGAUUGCAUACAAAUAUUUCUUUCAAUCAGUGGAAUUAUAGGAGUUUUGUGCGUCACAAAUCCGUGGUAUUUGAUUAAUACCAUC (((((...((((...)))))))))...(((((((..(((((.................))))).(..(((((.....)))))..))))))))......((((((((.....)))))))). ( -33.63) >DroSim_CAF1 3210 120 - 1 GGGCAAGUGGAUGAAAUCCUGCCCGCAGUGAUGCUCGAUUGCAUACAAAUAUUUCUGUCAAUCAGUGGAAUUAUAGGAGUUCUGUGCAUCACAAAUCCGUGGUAUUUGAUUAAUACCAUC (((((...((((...)))))))))...(((((((..(((((...(((........)))))))).(..(((((.....)))))..))))))))......((((((((.....)))))))). ( -37.80) >DroEre_CAF1 3004 120 - 1 GGGCAAGUGGACGAAAUCCUGCCCGCAGUGAUGCUCGAUUGCAUACAGAUAUUUCUCACAAUCAGUGGAAUUAUAGGGGUUUUGUGCAUCACCAAUCCCUGGUAUUUGAUUAAUACCAUC (((((...(((.....))))))))...(((((((.(((..((...(..(((.(((.(((.....)))))).)))..).)).))).))))))).......(((((((.....))))))).. ( -37.40) >DroYak_CAF1 3007 120 - 1 GGGCAAGUGGAUGAAAUCCUGCCCGCAGUGAUGCUAGAUUGCAUACAGAUAUUUCUCACAAUCAGUGGAAUUAUAGGAGUAUUGUGCAUCACAAAUCCCUGGUAUUUGAUUAAUACCAUC (((((...((((...)))))))))...(((((((.....(((...(..(((.(((.(((.....)))))).)))..).)))....))))))).......(((((((.....))))))).. ( -35.00) >consensus GGGCAAGUGGAUGAAAUCCUGCCCGCAGUGAUGCUCGAUUGCAUACAGAUAUUUCUCACAAUCAGUGGAAUUAUAGGAGUUUUGUGCAUCACAAAUCCGUGGUAUUUGAUUAAUACCAUC (((((...(((.....))))))))...(((((((..(((((.................))))).(..(((((.....)))))..)))))))).......(((((((.....))))))).. (-33.31 = -32.95 + -0.36)



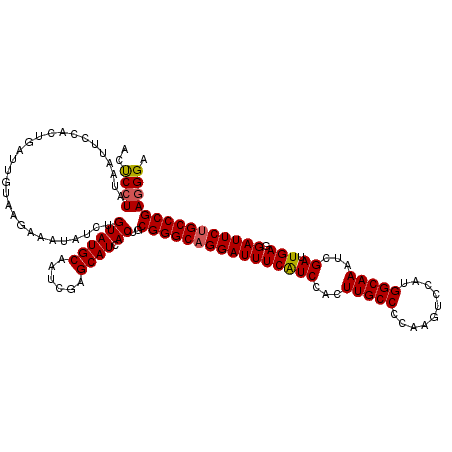
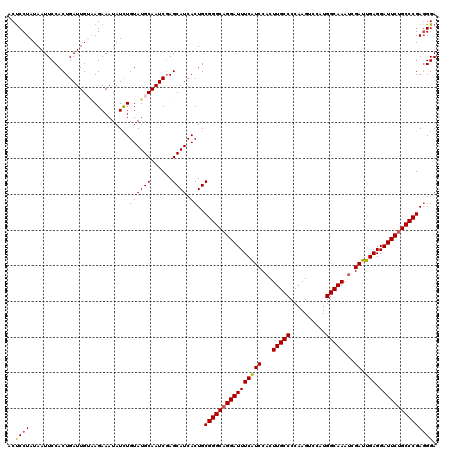
| Location | 20,918,636 – 20,918,756 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -30.78 |
| Energy contribution | -30.86 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20918636 120 + 22407834 ACUCCUAUAAUUCCACUGAUUGUAAGAAAUAUCUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCCCAAGUCCAUGGCAAAUCGAUUGAGGAUUCUGCCCGAUGGA ...........((((.(((((((((((....)))...)))))))).(((....)))(((((((((((((((...(((((..........)))))...)).))).))))))))))..)))) ( -35.40) >DroSec_CAF1 3990 120 + 1 ACUCCUAUAAUUCCACUGAUUGAAAGAAAUAUUUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCCCAAGUCCAUGGCAAAUCGAUUGAGGAUUCUGCCCGAGGGA ...........(((...................((.((((......))))))((.((((((((((((((((...(((((..........)))))...)).))).)))))))))))))))) ( -32.70) >DroSim_CAF1 3250 120 + 1 ACUCCUAUAAUUCCACUGAUUGACAGAAAUAUUUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCCCAAGUCCAUGGCAAAUCGAUUGAGGAUUCUGCCCGAGGGA ...........(((..(((((((((((....)))))...)))))).......((.((((((((((((((((...(((((..........)))))...)).))).)))))))))))))))) ( -34.60) >DroEre_CAF1 3044 120 + 1 ACCCCUAUAAUUCCACUGAUUGUGAGAAAUAUCUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCGUCCACUUGCCCCAAGUCCAUGGCAAAGCGAUUGAGGAUUCGGCCCGAGGGA ..((((....((((((.....))).)))......((((((......)))).))..(((((.((((((((..(.((((((..........))).))).)..))).))))).))))))))). ( -34.50) >DroYak_CAF1 3047 120 + 1 ACUCCUAUAAUUCCACUGAUUGUGAGAAAUAUCUGUAUGCAAUCUAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCCCAAGUCCAUGGCAAAUCGAUUGAGGAUUCUGCCCGAGGGA ..((((....((((((.....))).)))......((((((......)))).))..((((((((((((((((...(((((..........)))))...)).))).))))))))))))))). ( -36.00) >consensus ACUCCUAUAAUUCCACUGAUUGUAAGAAAUAUCUGUAUGCAAUCGAGCAUCACUGCGGGCAGGAUUUCAUCCACUUGCCCCAAGUCCAUGGCAAAUCGAUUGAGGAUUCUGCCCGAGGGA ..((((............................((((((......)))).))..((((((((((((((((...(((((..........)))))...)).))).))))))))))))))). (-30.78 = -30.86 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:21 2006