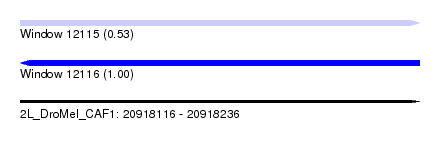
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,918,116 – 20,918,236 |
| Length | 120 |
| Max. P | 0.999689 |

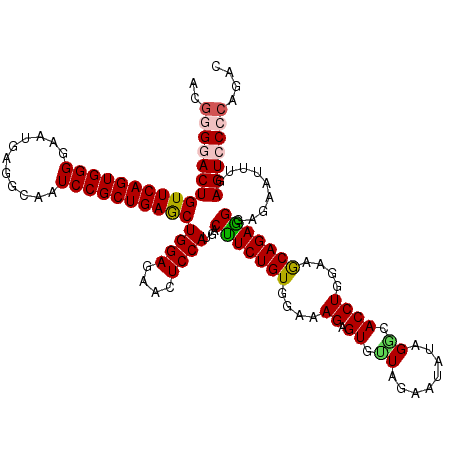
| Location | 20,918,116 – 20,918,236 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -22.66 |
| Energy contribution | -23.26 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20918116 120 + 22407834 GUCUGGGGACUCGAAUUCUCCCUCUGCUUCCAGGUGCCUAUAUUCUAGCACUCUUUCCACAGAAGUCAUGGAGUUCUCCAGCUCAGCGGAUUGCCUCAUUCCCCACUGAACAGUCCCCGU ....(((((((.(((((((.(.((((.....((((((..........)))).)).....)))).)....)))))))....(.((((.((.............)).)))).)))))))).. ( -32.82) >DroSec_CAF1 3470 120 + 1 GUCUGGGGACUCAAAUUCUCCCUCUGCUUCCAGGUGCCUAUAUUCUAAAACUCUUUCCACAGAGGUCAUGGAGUUCUCCAGCUCAGCGGAUUGCCGCAUUCCCCACUGAACAGUCCCCGU ....(((((((.........((((((.....((((..............)).)).....))))))(((((((((......)))).((((....))))......)).)))..))))))).. ( -33.64) >DroSim_CAF1 2730 120 + 1 CUCUGGGGACUCAAAUUCUUCCUCUGCUUCCAGGUGCCUAUAUUCUAACACUCUUUCCACAGAAGUCAUGGAGUUCUCCAGCUCAGCGGAUUGCCGCAUUCCCCACUGAACAGUCCCCGU ....(((((((........(((...(((((...(((.....................))).)))))...)))((((.........((((....))))..........))))))))))).. ( -31.01) >DroEre_CAF1 2524 120 + 1 GUCUGCGGACUCAAAUGCUCCUUCUGCUUCCAGGUUUCUGUAUUCCAAUACUCUUUCCACAGAAGUCAUGGAGUUCUCCAGCUCAGCGGAUUGCCUCAUGCCCCACUGAACAGUUCCCGU ....((((........(((((....(((((..((.....((((....)))).....))...)))))...)))))......(.((((.((...((.....)).)).)))).).....)))) ( -26.20) >DroYak_CAF1 2527 120 + 1 GCCUUCGGACUCAAAUACUCCUUCUGUUUCCAGGUUUCUAUAUUCUAUCACUCUUUCCACAGAAGUCAUGGAGUUCUCCAGUUCAGCGGAUUGCCUCAUUCCCCACUGAACAGUUCCCGU ......(((((.........(((((((....((((..............)).))....)))))))...((((....))))((((((.((.............)).))))))))))).... ( -23.16) >consensus GUCUGGGGACUCAAAUUCUCCCUCUGCUUCCAGGUGCCUAUAUUCUAACACUCUUUCCACAGAAGUCAUGGAGUUCUCCAGCUCAGCGGAUUGCCUCAUUCCCCACUGAACAGUCCCCGU ...((((((((.........((((((.....((((..............)).)).....))))))...((((....))))(.((((.((.............)).)))).))))))))). (-22.66 = -23.26 + 0.60)

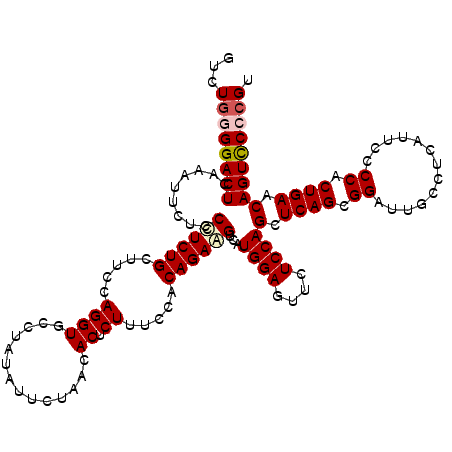
| Location | 20,918,116 – 20,918,236 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -36.76 |
| Energy contribution | -37.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20918116 120 - 22407834 ACGGGGACUGUUCAGUGGGGAAUGAGGCAAUCCGCUGAGCUGGAGAACUCCAUGACUUCUGUGGAAAGAGUGCUAGAAUAUAGGCACCUGGAAGCAGAGGGAGAAUUCGAGUCCCCAGAC ..(((((((((((((((((...........))))))))))((((....))))...(((((((....((.(((((........)))))))....))))))).........))))))).... ( -50.10) >DroSec_CAF1 3470 120 - 1 ACGGGGACUGUUCAGUGGGGAAUGCGGCAAUCCGCUGAGCUGGAGAACUCCAUGACCUCUGUGGAAAGAGUUUUAGAAUAUAGGCACCUGGAAGCAGAGGGAGAAUUUGAGUCCCCAGAC ..(((((((((((((((((...........))))))))))((((....))))...(((((((....((.((((((.....)))).))))....))))))).........))))))).... ( -45.70) >DroSim_CAF1 2730 120 - 1 ACGGGGACUGUUCAGUGGGGAAUGCGGCAAUCCGCUGAGCUGGAGAACUCCAUGACUUCUGUGGAAAGAGUGUUAGAAUAUAGGCACCUGGAAGCAGAGGAAGAAUUUGAGUCCCCAGAG ..(((((((((((((((((...........))))))))))((((....))))...(((((((....((.(((((........)))))))....))))))).........))))))).... ( -47.40) >DroEre_CAF1 2524 120 - 1 ACGGGAACUGUUCAGUGGGGCAUGAGGCAAUCCGCUGAGCUGGAGAACUCCAUGACUUCUGUGGAAAGAGUAUUGGAAUACAGAAACCUGGAAGCAGAAGGAGCAUUUGAGUCCGCAGAC .........(((((((((.((.....))...)))))))))((((....)))).....((((((((..((((.(((.....)))....(((....))).......))))...)))))))). ( -36.40) >DroYak_CAF1 2527 120 - 1 ACGGGAACUGUUCAGUGGGGAAUGAGGCAAUCCGCUGAACUGGAGAACUCCAUGACUUCUGUGGAAAGAGUGAUAGAAUAUAGAAACCUGGAAACAGAAGGAGUAUUUGAGUCCGAAGGC .((((....((((((((((...........))))))))))(.(((.(((((.....(((((((.....(....)....)))))))..(((....)))..))))).))).).))))..... ( -36.40) >consensus ACGGGGACUGUUCAGUGGGGAAUGAGGCAAUCCGCUGAGCUGGAGAACUCCAUGACUUCUGUGGAAAGAGUGUUAGAAUAUAGGCACCUGGAAGCAGAGGGAGAAUUUGAGUCCCCAGAC ..(((((((((((((((((...........))))))))))((((....))))...(((((((....((.((.((........)).))))....))))))).........))))))).... (-36.76 = -37.00 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:16 2006