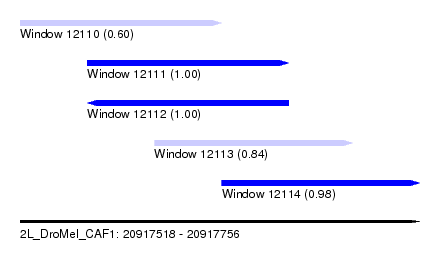
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,917,518 – 20,917,756 |
| Length | 238 |
| Max. P | 0.999120 |

| Location | 20,917,518 – 20,917,638 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -38.69 |
| Consensus MFE | -34.38 |
| Energy contribution | -34.26 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20917518 120 + 22407834 UGCGGUAGCCUCGUCCAUCACAAGGAUGCGGUUCUCGCGGAGAAUCGCCCUGGCCAAGCAGACCAACUGCCUCUGACCUACACUGUAGUUGGAUCCGCCCUCGGCAACCACACUUUCCAG .(((..((((.(((((.......))))).))))..)))(((((.........(((..((.((((((((((...((.....))..)))))))).)).))....)))........))))).. ( -40.53) >DroSec_CAF1 2876 120 + 1 UGCGGUAGCCUCGUCCAUCACAAGGAUGCGGUUCUCGCGGAGAAUCGCCCUGGCCAAGCAGACCAGCUGCCUCUGACCUACACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAG .(((..((((.(((((.......))))).))))..)))(((((.........(((..((.((((((((((...((.....))..)))))))).)).))....)))........))))).. ( -40.53) >DroSim_CAF1 2136 120 + 1 UGCGGUAGUCUCGUCCAUCACCAGGAUGCGGUUCUCGCGGAGAAUCGCCCUGGCCAAGCAGACCAACUGCCUCUGACCUACACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAG ((.(((.(((..........(((((..(((((((((...))))))))))))))....((.((((((((((...((.....))..)))))))).)).))....))).))).))........ ( -41.00) >DroEre_CAF1 1927 120 + 1 GGCAGUAGCCUCGUCCAUGACAAGGAUGCGGUUCUCGCGGAGGAUCGCCCUGGCCAAGCAGACCAAUUGCCUCUGACCUACACUGUAGUUGGCUCCGCCUUCUGCAACCACACUUUCCAA .((((.((((.(((((.......))))).))))...((((((....((....))........((((((((...((.....))..))))))))))))))...))))............... ( -36.40) >DroYak_CAF1 1931 120 + 1 CGCGGUAGCCUCGUCCAUCACAAGGAUACGGUUCUCGCGGAGAAUCGCCCUGGCCAAGCAGACCAAUUGCCUCUGACCUACACUGUAGUUGGCUCCACCCUCUGCAACCACACUUUCCAG .(((..((((..((((.......))))..))))..)))(((((...((....))...(((((((((((((...((.....))..))))))))........)))))........))))).. ( -35.00) >consensus UGCGGUAGCCUCGUCCAUCACAAGGAUGCGGUUCUCGCGGAGAAUCGCCCUGGCCAAGCAGACCAACUGCCUCUGACCUACACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAG .((((.((((.(((((.......))))).)))).))))(((((...((....))...((.((((((((((...((.....))..)))))))).)).))...............))))).. (-34.38 = -34.26 + -0.12)


| Location | 20,917,558 – 20,917,678 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -33.42 |
| Energy contribution | -33.22 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20917558 120 + 22407834 AGAAUCGCCCUGGCCAAGCAGACCAACUGCCUCUGACCUACACUGUAGUUGGAUCCGCCCUCGGCAACCACACUUUCCAGGCCAUUCGGCAGCUCGGAUACCUCAUCUUUGAGGUGCACC ......(((.(((((..((.((((((((((...((.....))..)))))))).)).))....(....)...........)))))...))).....(..(((((((....)))))))..). ( -38.40) >DroSec_CAF1 2916 120 + 1 AGAAUCGCCCUGGCCAAGCAGACCAGCUGCCUCUGACCUACACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAGGCCAUUUGGCAGCUUCGACACCUCAUCUUUGAGGUGAACC .(((.((((.(((((..(.((.((((((((...((.....))..)))))))))).)(((...)))..............)))))...))).).)))..(((((((....))))))).... ( -40.90) >DroSim_CAF1 2176 120 + 1 AGAAUCGCCCUGGCCAAGCAGACCAACUGCCUCUGACCUACACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAGGCCAUUUGGCAGCUCCGACACCUCAUCUUUGAGGUGAACC ......(((.(((((..(.((.((((((((...((.....))..)))))))))).)(((...)))..............)))))...)))........(((((((....))))))).... ( -40.20) >DroEre_CAF1 1967 120 + 1 AGGAUCGCCCUGGCCAAGCAGACCAAUUGCCUCUGACCUACACUGUAGUUGGCUCCGCCUUCUGCAACCACACUUUCCAAGCCAUUUGGCAGCUCGGAUACCUCAUCCUUGAGGUGAACC .((.((..((.(((...(((((((((((((...((.....))..))))))))........)))))...............(((....))).))).))..((((((....)))))))).)) ( -33.30) >DroYak_CAF1 1971 120 + 1 AGAAUCGCCCUGGCCAAGCAGACCAAUUGCCUCUGACCUACACUGUAGUUGGCUCCACCCUCUGCAACCACACUUUCCAGGCCUUUUGGCAGCUCGGACACUUCAUCUUUGAGGUGAACC ........((.(((...(((((((((((((...((.....))..))))))))........)))))...............(((....))).))).)).(((((((....))))))).... ( -32.00) >consensus AGAAUCGCCCUGGCCAAGCAGACCAACUGCCUCUGACCUACACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAGGCCAUUUGGCAGCUCGGACACCUCAUCUUUGAGGUGAACC ......(((.(((((.....((((((((((...((.....))..)))))))).)).((.....))..............)))))...)))........(((((((....))))))).... (-33.42 = -33.22 + -0.20)

| Location | 20,917,558 – 20,917,678 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -47.02 |
| Consensus MFE | -42.16 |
| Energy contribution | -43.08 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20917558 120 - 22407834 GGUGCACCUCAAAGAUGAGGUAUCCGAGCUGCCGAAUGGCCUGGAAAGUGUGGUUGCCGAGGGCGGAUCCAACUACAGUGUAGGUCAGAGGCAGUUGGUCUGCUUGGCCAGGGCGAUUCU ..(((((((((....))))))..((.(((((((...(((((((.(...(((((((((((....)))...)))))))).).)))))))..)))))))((((.....)))).)))))..... ( -51.60) >DroSec_CAF1 2916 120 - 1 GGUUCACCUCAAAGAUGAGGUGUCGAAGCUGCCAAAUGGCCUGGAAAGUGUGGUUGCCGAGGGCGGAGCCAACUACAGUGUAGGUCAGAGGCAGCUGGUCUGCUUGGCCAGGGCGAUUCU ....(((((((....)))))))(((.(((((((...(((((((.(...(((((((((((....)))...)))))))).).)))))))..)))))))((((.....))))....))).... ( -52.50) >DroSim_CAF1 2176 120 - 1 GGUUCACCUCAAAGAUGAGGUGUCGGAGCUGCCAAAUGGCCUGGAAAGUGUGGUUGCCGAGGGCGGAGCCAACUACAGUGUAGGUCAGAGGCAGUUGGUCUGCUUGGCCAGGGCGAUUCU ....(((((((....)))))))(((.(((((((...(((((((.(...(((((((((((....)))...)))))))).).)))))))..)))))))((((.....))))....))).... ( -50.40) >DroEre_CAF1 1967 120 - 1 GGUUCACCUCAAGGAUGAGGUAUCCGAGCUGCCAAAUGGCUUGGAAAGUGUGGUUGCAGAAGGCGGAGCCAACUACAGUGUAGGUCAGAGGCAAUUGGUCUGCUUGGCCAGGGCGAUCCU ((.((((((((....))))))......((((((...(((((((.(...(((((((((..........).)))))))).).)))))))..)))..((((((.....))))))))))).)). ( -41.90) >DroYak_CAF1 1971 120 - 1 GGUUCACCUCAAAGAUGAAGUGUCCGAGCUGCCAAAAGGCCUGGAAAGUGUGGUUGCAGAGGGUGGAGCCAACUACAGUGUAGGUCAGAGGCAAUUGGUCUGCUUGGCCAGGGCGAUUCU ((..(((.(((....))).))).))(((.((((....((((((.(..((((((((.((.....)).)))))..)))..).))))))........((((((.....)))))))))).))). ( -38.70) >consensus GGUUCACCUCAAAGAUGAGGUGUCCGAGCUGCCAAAUGGCCUGGAAAGUGUGGUUGCCGAGGGCGGAGCCAACUACAGUGUAGGUCAGAGGCAGUUGGUCUGCUUGGCCAGGGCGAUUCU .((((((((((....)))))).....(((((((...(((((((.(...(((((((((((....)))...)))))))).).)))))))..)))))))((((.....)))).))))...... (-42.16 = -43.08 + 0.92)

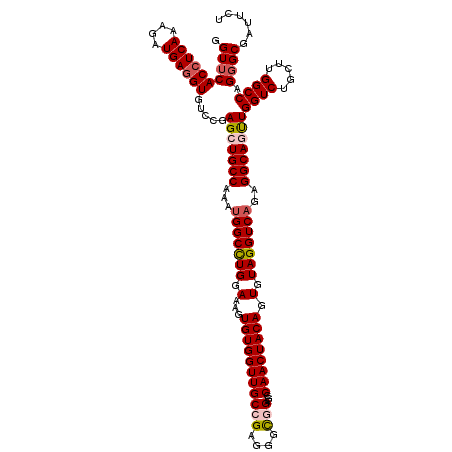

| Location | 20,917,598 – 20,917,716 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -27.12 |
| Energy contribution | -29.08 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20917598 118 + 22407834 CACUGUAGUUGGAUCCGCCCUCGGCAACCACACUUUCCAGGCCAUUCGGCAGCUCGGAUACCUCAUCUUUGAGGUGCACCUGGCGAGGGAAAAUGGUUUAAUUAUAGUU--AUGAUUGUC .((((((((((((((..((((((.((.........((((((((....)))..)).)))(((((((....)))))))....)).)))))).....)))))))))))))).--......... ( -42.50) >DroSec_CAF1 2956 115 + 1 CACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAGGCCAUUUGGCAGCUUCGACACCUCAUCUUUGAGGUGAACCUGGCGAGAAAAGGGUGAUUAGUUAUAGUU--AUG---GUU .(((((((.(((((.((((((.(....)....(((.(((((((....)))........(((((((....)))))))...)))).)))...))))))...)))))...))--)))---)). ( -39.10) >DroSim_CAF1 2216 115 + 1 CACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAGGCCAUUUGGCAGCUCCGACACCUCAUCUUUGAGGUGAACCUGGCGAGGAAAGGGUGAUUAGUUAUAGUU--AUG---GUC .(((((((.(((((.((((((.(....)....(((.(((((((....)))........(((((((....)))))))...)))).)))...))))))...)))))...))--)))---)). ( -39.30) >DroEre_CAF1 2007 118 + 1 CACUGUAGUUGGCUCCGCCUUCUGCAACCACACUUUCCAAGCCAUUUGGCAGCUCGGAUACCUCAUCCUUGAGGUGAACCUGACGAAGAAAGAGUGUUCAUUUAUAAUU--AUAGCUUUU ...(((((..(((...)))..)))))..(((.(((((...(((....))).(.((((.(((((((....)))))))...)))))...))))).))).............--......... ( -32.20) >DroYak_CAF1 2011 119 + 1 CACUGUAGUUGGCUCCACCCUCUGCAACCACACUUUCCAGGCCUUUUGGCAGCUCGGACACUUCAUCUUUGAGGUGAACCUGGCGAAGGAAGAGUGUUUAUUUG-AAUUAAAUAACUUUU ...(((((..((.....))..)))))...(((((((((..(((....))).(((.((.(((((((....)))))))..)).)))...)).)))))))(((((((-...)))))))..... ( -35.40) >consensus CACUGUAGUUGGCUCCGCCCUCGGCAACCACACUUUCCAGGCCAUUUGGCAGCUCGGACACCUCAUCUUUGAGGUGAACCUGGCGAGGAAAGAGUGUUUAGUUAUAGUU__AUG__UGUU .(((((((((((....((.....))...(((.((((((..(((....))).(((.((.(((((((....)))))))..)).)))..)))))).))).)))))))))))............ (-27.12 = -29.08 + 1.96)



| Location | 20,917,638 – 20,917,756 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -36.26 |
| Consensus MFE | -23.40 |
| Energy contribution | -22.44 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20917638 118 + 22407834 GCCAUUCGGCAGCUCGGAUACCUCAUCUUUGAGGUGCACCUGGCGAGGGAAAAUGGUUUAAUUAUAGUU--AUGAUUGUCAGGUUUGGUAAUUACUCACCUCUUCUAAGGCCUCCCACAA (((....))).(((.((.(((((((....)))))))..)).)))..((((....(((((((((((....--))))))...((((..(((....))).))))......))))))))).... ( -32.90) >DroSec_CAF1 2996 114 + 1 GCCAUUUGGCAGCUUCGACACCUCAUCUUUGAGGUGAACCUGGCGAGAAAAGGGUGAUUAGUUAUAGUU--AUG---GUUAGGUUU-GUAAUUACUCACCUCUUCUAAGGCCUUCCACAA (((....)))........(((((((....))))))).....(((.((((..((((((.(((((((((..--...---.......))-))))))).)))))).))))...)))........ ( -36.40) >DroSim_CAF1 2256 114 + 1 GCCAUUUGGCAGCUCCGACACCUCAUCUUUGAGGUGAACCUGGCGAGGAAAGGGUGAUUAGUUAUAGUU--AUG---GUCAGGUUU-GUAAUUUCUCACCUCUUCUAAGGCCUUCCACAA (((....)))........(((((((....))))))).....(((..((((..(((((..((((((((..--...---.......))-))))))..)))))..))))...)))........ ( -37.00) >DroEre_CAF1 2047 117 + 1 GCCAUUUGGCAGCUCGGAUACCUCAUCCUUGAGGUGAACCUGACGAAGAAAGAGUGUUCAUUUAUAAUU--AUAGCUUUUGGACGU-GGAGACUCUCACUUCUUCCAGGGCCUCCCACAA (((....))).....((((((((((....)))))))..((((..(((((((((((.(((((...(((..--.......)))...))-))).)))))...))))))))))...)))..... ( -37.00) >DroYak_CAF1 2051 116 + 1 GCCUUUUGGCAGCUCGGACACUUCAUCUUUGAGGUGAACCUGGCGAAGGAAGAGUGUUUAUUUG-AAUUAAAUAACUUUUGG--GU-AAAAACUCUCACUUCUUCUAGUGCCUCCCACAA (((....))).(((.((.(((((((....)))))))..)).)))((((..(((((.((((((..-((..........))..)--))-))).)))))..)))).....(((.....))).. ( -38.00) >consensus GCCAUUUGGCAGCUCGGACACCUCAUCUUUGAGGUGAACCUGGCGAGGAAAGAGUGUUUAGUUAUAGUU__AUG__UGUUAGGUUU_GUAAUUUCUCACCUCUUCUAAGGCCUCCCACAA (((.((((.(((......(((((((....)))))))...))).))))(((.(((((...((((((.......((.....))......))))))...))))).)))...)))......... (-23.40 = -22.44 + -0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:14 2006