
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,917,239 – 20,917,398 |
| Length | 159 |
| Max. P | 0.983600 |

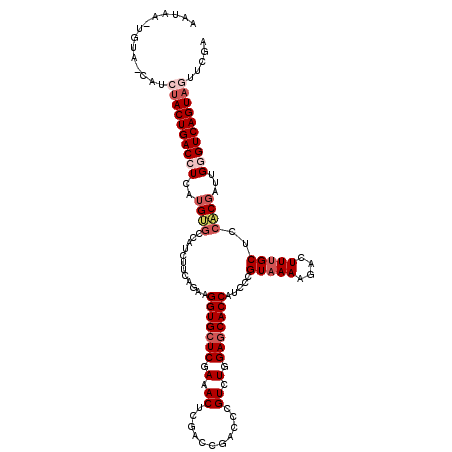
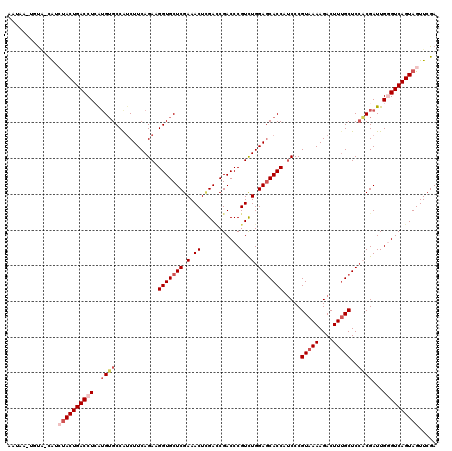
| Location | 20,917,239 – 20,917,358 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -25.62 |
| Energy contribution | -27.38 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20917239 119 + 22407834 AAUAAAUAUU-CAUCUACUGACCUCAAGUGCCAGCUUCAGAAGGUGCUCGAAACUCGACCGACCCGUCUGGAGCACCAUCCCGUAAAAGACUUUGCUCCACGAUUGGGUCAGUAGUUCGA ..........-...((((((((((...(((...((((((((.((((.(((.....))).).)))..))))))))........(((((....)))))..)))....))))))))))..... ( -38.70) >DroSec_CAF1 2597 119 + 1 AAUAAGAGUA-CAUCUACUGACCUCAUGUGCCGUCUUCAGAAGGUGCUCGAAACUCGAACGACCCGUUUGGAGCACCAACCCGUAAAAGACUUUGCUCCACGAUUGGGUCAGUAGUUCGA ..........-...((((((((((.(((((............(((((((.((((((....))...)))).))))))).....(((((....)))))..))).)).))))))))))..... ( -37.80) >DroSim_CAF1 1857 119 + 1 AAUAAAAGUA-CAUCUACUGACCUCAUGUGCCGUCUUCAGAAGGUGCUCGAAACUCGACCGACCCGUCUGGAGCACCAUCCCGUAAAAGACUUUGCUCCACGAUUGGGUCAGUAGUUCGA ..........-...((((((((((...(..((.((....)).))..)(((......(((......)))(((((((...((........))...))))))))))..))))))))))..... ( -36.00) >DroEre_CAF1 1650 117 + 1 --UAU-UUUACUGUACACUGACCUGGUGGGCCAUCUUCAGAAGGUGCUCGAAACUUGAUCGACCCGUCUGGAACACCAUCCCGUAAAAGACUUUGCUCGCCGAUUGCGUCAGUAGUUCGA --...-.....((.((((((((.((((.(((....((((((.((.(.((((.......))))))).))))))..........(((((....)))))..))).)))).)))))).)).)). ( -30.10) >DroYak_CAF1 1653 118 + 1 AAUAA-UCCG-CGUAUACUGACCUGAUGUGCCAUCUUCAGAAGGUGCUCGAAACUGGACCGACCCGUCUGGAGCACCAUCCCGUAAAAGACUUGGCUCCCCGAUUGCGUCAGUAGUUCGA .....-....-((..(((((((.(((((.((((((((..((.(((((((.(.((.((.....)).)).).))))))).))......))))..)))).)....)))).)))))))...)). ( -34.10) >consensus AAUAA_UGUA_CAUCUACUGACCUCAUGUGCCAUCUUCAGAAGGUGCUCGAAACUCGACCGACCCGUCUGGAGCACCAUCCCGUAAAAGACUUUGCUCCACGAUUGGGUCAGUAGUUCGA ..............((((((((((..((((............(((((((.(.((...........)).).))))))).....(((((....)))))..))))...))))))))))..... (-25.62 = -27.38 + 1.76)

| Location | 20,917,239 – 20,917,358 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -43.48 |
| Consensus MFE | -33.70 |
| Energy contribution | -34.42 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20917239 119 - 22407834 UCGAACUACUGACCCAAUCGUGGAGCAAAGUCUUUUACGGGAUGGUGCUCCAGACGGGUCGGUCGAGUUUCGAGCACCUUCUGAAGCUGGCACUUGAGGUCAGUAGAUG-AAUAUUUAUU .....(((((((((...((.(((((((..((((......))))..)))))))))(((((..(((.(((((((((....))).)))))))))))))).)))))))))...-.......... ( -44.20) >DroSec_CAF1 2597 119 - 1 UCGAACUACUGACCCAAUCGUGGAGCAAAGUCUUUUACGGGUUGGUGCUCCAAACGGGUCGUUCGAGUUUCGAGCACCUUCUGAAGACGGCACAUGAGGUCAGUAGAUG-UACUCUUAUU .....(((((((((...(((((..((...(((((...((((..(((((((.((((.((....))..)))).))))))).))))))))).)).))))))))))))))...-.......... ( -46.50) >DroSim_CAF1 1857 119 - 1 UCGAACUACUGACCCAAUCGUGGAGCAAAGUCUUUUACGGGAUGGUGCUCCAGACGGGUCGGUCGAGUUUCGAGCACCUUCUGAAGACGGCACAUGAGGUCAGUAGAUG-UACUUUUAUU .....(((((((((...(((((..((...(((((...(((((.(((((((.((((..(.....)..)))).))))))))))))))))).)).))))))))))))))...-.......... ( -47.90) >DroEre_CAF1 1650 117 - 1 UCGAACUACUGACGCAAUCGGCGAGCAAAGUCUUUUACGGGAUGGUGUUCCAGACGGGUCGAUCAAGUUUCGAGCACCUUCUGAAGAUGGCCCACCAGGUCAGUGUACAGUAAA-AUA-- ......(((((((((.(((((.(.((...(((((...(((((.(((((((.((((..(.....)..)))).))))))))))))))))).)).).)).)))..)))).)))))..-...-- ( -37.40) >DroYak_CAF1 1653 118 - 1 UCGAACUACUGACGCAAUCGGGGAGCCAAGUCUUUUACGGGAUGGUGCUCCAGACGGGUCGGUCCAGUUUCGAGCACCUUCUGAAGAUGGCACAUCAGGUCAGUAUACG-CGGA-UUAUU (((...(((((((......((....))..(((((((.(((((.(((((((.((((.((.....)).)))).)))))))))))))))).))).......)))))))....-))).-..... ( -41.40) >consensus UCGAACUACUGACCCAAUCGUGGAGCAAAGUCUUUUACGGGAUGGUGCUCCAGACGGGUCGGUCGAGUUUCGAGCACCUUCUGAAGAUGGCACAUGAGGUCAGUAGAUG_UACA_UUAUU .....(((((((((...(((....((...(((((...(((((.(((((((.((((..(.....)..)))).))))))))))))))))).))...)))))))))))).............. (-33.70 = -34.42 + 0.72)

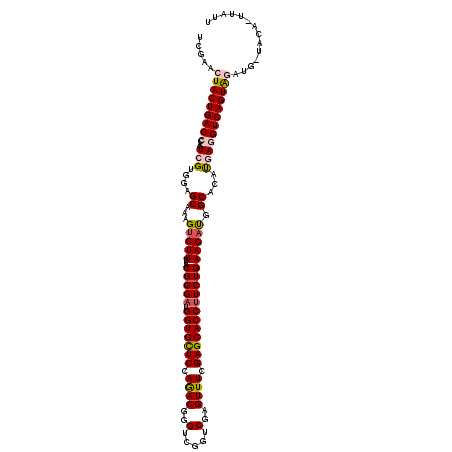
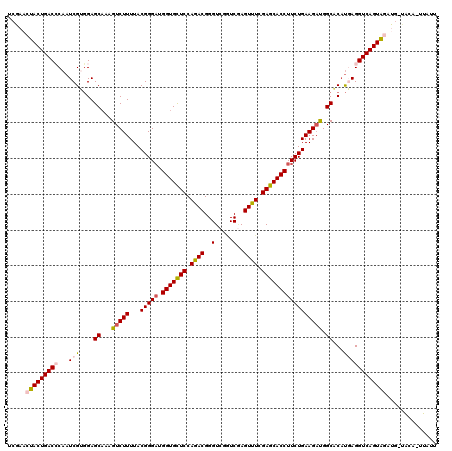
| Location | 20,917,278 – 20,917,398 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -45.54 |
| Consensus MFE | -37.92 |
| Energy contribution | -39.00 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20917278 120 - 22407834 GUUCUGGACGCUGGUACUCUAGUGGAGUUCGGAUCACCAUUCGAACUACUGACCCAAUCGUGGAGCAAAGUCUUUUACGGGAUGGUGCUCCAGACGGGUCGGUCGAGUUUCGAGCACCUU ((((..((((((((....)))))..(((((((((....)))))))))((((((((..((.(((((((..((((......))))..))))))))).))))))))...)))..))))..... ( -50.70) >DroSec_CAF1 2636 120 - 1 GUUCUGGACGCUGGCACUCUAGUGGAGUUUGGGUCACCAUUCGAACUACUGACCCAAUCGUGGAGCAAAGUCUUUUACGGGUUGGUGCUCCAAACGGGUCGUUCGAGUUUCGAGCACCUU ((((..(((.(..(.((((.....)))))..))))...((((((((....(((((.....(((((((.(..((.....))..)..)))))))...)))))))))))))...))))..... ( -42.90) >DroSim_CAF1 1896 120 - 1 GUUCUGGACGCUGGCACUCUAGUGGAGUUCGGGUCACCAUUCGAACUACUGACCCAAUCGUGGAGCAAAGUCUUUUACGGGAUGGUGCUCCAGACGGGUCGGUCGAGUUUCGAGCACCUU ((((..((((((((....)))))..(((((((((....)))))))))((((((((..((.(((((((..((((......))))..))))))))).))))))))...)))..))))..... ( -49.20) >DroEre_CAF1 1687 120 - 1 GUUCUGGACGCUGGCACUCUGGUGGAGUUCGGGUCUCCAUUCGAACUACUGACGCAAUCGGCGAGCAAAGUCUUUUACGGGAUGGUGUUCCAGACGGGUCGAUCAAGUUUCGAGCACCUU ((...(((((((.((......(((.(((((((((....))))))))).....))).....)).)))...))))...)).....(((((((.((((..(.....)..)))).))))))).. ( -37.60) >DroYak_CAF1 1691 120 - 1 GUUCUGGACGCUGGCACUCUGGUAGAGUUCGGGUCUCCAUUCGAACUACUGACGCAAUCGGGGAGCCAAGUCUUUUACGGGAUGGUGCUCCAGACGGGUCGGUCCAGUUUCGAGCACCUU ((...((((..((((.((((((((((((((((((....))))))))).))......))))))).)))).))))...)).....(((((((.((((.((.....)).)))).))))))).. ( -47.30) >consensus GUUCUGGACGCUGGCACUCUAGUGGAGUUCGGGUCACCAUUCGAACUACUGACCCAAUCGUGGAGCAAAGUCUUUUACGGGAUGGUGCUCCAGACGGGUCGGUCGAGUUUCGAGCACCUU ((((..((((((((....)))))..(((((((((....)))))))))((((((((.....(((((((..((((......))))..)))))))...))))))))...)))..))))..... (-37.92 = -39.00 + 1.08)

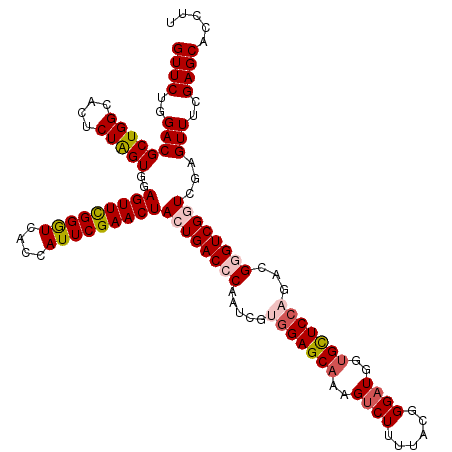
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:10 2006