
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,893,665 – 20,893,756 |
| Length | 91 |
| Max. P | 0.669236 |

| Location | 20,893,665 – 20,893,756 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -9.11 |
| Energy contribution | -10.69 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
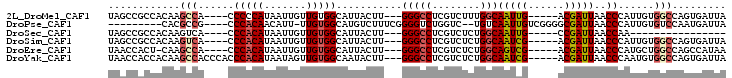
>2L_DroMel_CAF1 20893665 91 + 22407834 UAAUCACUGGCCACAAUGGGUUAAUCGU-----CAAUUGCCAAAGACGAGGCCC---AAGUAAUGCCACAACAAUUAUGGGGG----UGGCUUGUGGCGGCUA .........((((((((((((...((((-----(..........))))).))))---)......(((((..(.......)..)----)))))))))))..... ( -33.80) >DroPse_CAF1 86500 87 + 1 UAAUCAUUGGACACAAUGGGUUAAUCGCCCCGACAAUUGACA--GACCAGACCCCGAAAGACAUGCCACAA-AAUGUUGUGGG----CGGCGUG--------- ...((((((....)))))).....(((((((((((.(((...--...)))....(....)...........-..))))).)))----)))....--------- ( -22.20) >DroSec_CAF1 76809 75 + 1 ----------------UUGGUUAAUCGG-----CAAUUGCCAGAGACGAGGCCC---AAGUAAUGCCACAACAAUUAUGUGGG----UGACUUGUGGCGGCUA ----------------.(((((....((-----(....))).........((((---((((....(((((.......))))).----..))))).)))))))) ( -23.60) >DroSim_CAF1 80894 91 + 1 UAAUCACUGGCCACAAUGGGUUAAUCGU-----CGAUUGCCAGAGACGAGGCCC---AAGUAAUGCCACAACAAUUAUGUGGG----UGACUUGUGGCGGCUA .......(((((....(((((...((((-----(..........))))).))))---)......(((((((((....)))((.----...))))))))))))) ( -32.60) >DroEre_CAF1 78541 90 + 1 UUAUGGCUGGCCAGCAUGGGUUAAUCGU-----CGACUGCCAGAGACGAGGCCC---AAGUAAUGCCACAACAAUUAUGUGGG----UGGCUUG-AGUGGUUA .......((((((((.(((((...((((-----(..........))))).))))---).))...(((((.(((....)))..)----))))...-..)))))) ( -30.10) >DroYak_CAF1 69629 95 + 1 UAAUCACUGGCCACAUUGGGUUAAUCGU-----CGAUUGCCAGAGACGAGGCCC---AAGUAUUGCCACAACUAUUAUGUGGGUGGGUGGCUUGUGGUGGUUA ((((((((((((((.((((((...((((-----(..........))))).))))---))......((((.((......))..))))))))))...)))))))) ( -38.50) >consensus UAAUCACUGGCCACAAUGGGUUAAUCGU_____CAAUUGCCAGAGACGAGGCCC___AAGUAAUGCCACAACAAUUAUGUGGG____UGGCUUGUGGCGGCUA ........((((.....((((...(((...................))).))))...........(((((.......)))))................)))). ( -9.11 = -10.69 + 1.58)



| Location | 20,893,665 – 20,893,756 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -12.09 |
| Energy contribution | -12.62 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
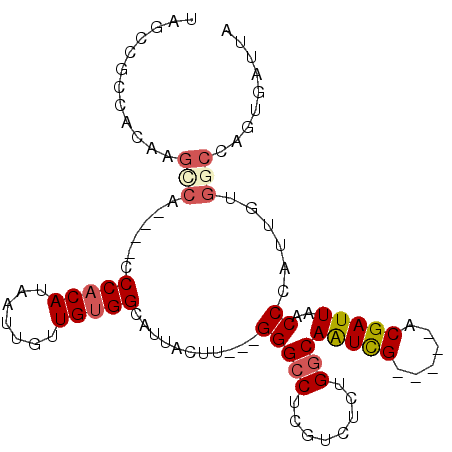
>2L_DroMel_CAF1 20893665 91 - 22407834 UAGCCGCCACAAGCCA----CCCCCAUAAUUGUUGUGGCAUUACUU---GGGCCUCGUCUUUGGCAAUUG-----ACGAUUAACCCAUUGUGGCCAGUGAUUA .....(((((((((((----(...((....))..)))))......(---(((..(((((..(....)..)-----))))....)))))))))))......... ( -29.70) >DroPse_CAF1 86500 87 - 1 ---------CACGCCG----CCCACAACAUU-UUGUGGCAUGUCUUUCGGGGUCUGGUC--UGUCAAUUGUCGGGGCGAUUAACCCAUUGUGUCCAAUGAUUA ---------..(((((----((.((((....-))))))).......((((.(..(((..--..)))..).)))))))).......(((((....))))).... ( -20.80) >DroSec_CAF1 76809 75 - 1 UAGCCGCCACAAGUCA----CCCACAUAAUUGUUGUGGCAUUACUU---GGGCCUCGUCUCUGGCAAUUG-----CCGAUUAACCAA---------------- .....(((.(((((..----.(((((.......)))))....))))---))))........((((....)-----))).........---------------- ( -20.80) >DroSim_CAF1 80894 91 - 1 UAGCCGCCACAAGUCA----CCCACAUAAUUGUUGUGGCAUUACUU---GGGCCUCGUCUCUGGCAAUCG-----ACGAUUAACCCAUUGUGGCCAGUGAUUA .....(((.(((((..----.(((((.......)))))....))))---))))...(((.(((((.....-----(((((......))))).))))).))).. ( -28.10) >DroEre_CAF1 78541 90 - 1 UAACCACU-CAAGCCA----CCCACAUAAUUGUUGUGGCAUUACUU---GGGCCUCGUCUCUGGCAGUCG-----ACGAUUAACCCAUGCUGGCCAGCCAUAA ........-...((((----(..(((....))).)))))......(---(((..(((((..........)-----))))....))))...(((....)))... ( -21.70) >DroYak_CAF1 69629 95 - 1 UAACCACCACAAGCCACCCACCCACAUAAUAGUUGUGGCAAUACUU---GGGCCUCGUCUCUGGCAAUCG-----ACGAUUAACCCAAUGUGGCCAGUGAUUA .......(((..(((((....(((((.......)))))......((---(((..(((((..........)-----))))....))))).)))))..))).... ( -27.70) >consensus UAGCCGCCACAAGCCA____CCCACAUAAUUGUUGUGGCAUUACUU___GGGCCUCGUCUCUGGCAAUCG_____ACGAUUAACCCAUUGUGGCCAGUGAUUA ............(((......(((((.......)))))...........(((((........)))(((((......)))))..))......)))......... (-12.09 = -12.62 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:00 2006