
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,892,161 – 20,892,258 |
| Length | 97 |
| Max. P | 0.832749 |

| Location | 20,892,161 – 20,892,258 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -25.27 |
| Energy contribution | -26.61 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
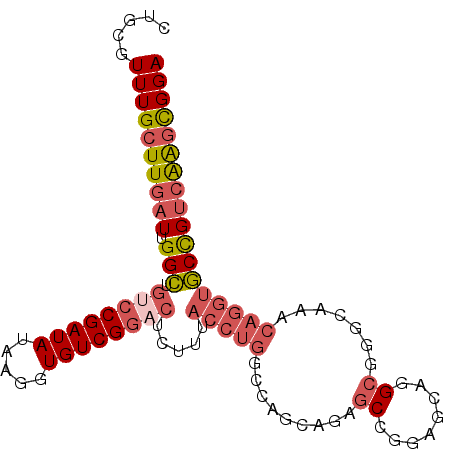
>2L_DroMel_CAF1 20892161 97 + 22407834 CUGCGUUUGCUUGAUUGGCUGUCCGAUAUAAGGUGUCGGACUUUUUACCUGGCCAGCAGAGCCAGAGCAGGCGGGGAAACAGGUGCCGUCAAGCGGA .....((((((((((.(((.(((((((((...))))))))).....(((((.((......(((......)))..))...)))))))))))))))))) ( -41.20) >DroSec_CAF1 75599 97 + 1 CUGCGUUUGCUUGAUUGGCUGUCCGAUAUAAGGUGUCGGACUCUUUACCUGGCCAGCAGAGCCGGAGCAGGCGGGCAAACAGGUGCCGUCAAGCGGA .....((((((((((.(((.(((((((((...))))))))).....(((((....((...(((......)))..))...)))))))))))))))))) ( -42.90) >DroSim_CAF1 79341 97 + 1 CUGCGUUUGCUUGAUUGGCUGUCCGAUAUAAGGUGUCGGACUCUUUACCUGGCCAGCAGAGCCGGAGCAGGCGGGCAAACAGGUGCCGUCAAGCGGA .....((((((((((.(((.(((((((((...))))))))).....(((((....((...(((......)))..))...)))))))))))))))))) ( -42.90) >DroEre_CAF1 77090 92 + 1 CUGCGUUUGCUUGAUUGGCUGUCCGAUAUAAGGUGUCGGUCUCUUUACCUGGCCAGCAGAGCAGU-----GCGGGGAAACAGGUGCCGUGGAGCGGA .....(((((((.((.(((.(.(((((((...))))))).).....(((((.((.(((......)-----))..))...)))))))))).))))))) ( -32.80) >DroYak_CAF1 68143 92 + 1 CUGCGUUUGCUUGAUUGGCUGUCCGAUAUAAGGUGUCGGACUUUUUACCUGG-CAGCAGAGCAGUA----GCGGGGAAACAGGUGCCGUCAAGCGGA .....((((((((((.(((.(((((((((...))))))))).....(((((.-...(...((....----))...)...)))))))))))))))))) ( -36.70) >DroAna_CAF1 63952 75 + 1 CUGCGUUUGCUUGAUUGGUUGUCCGAUAGAAGGUGUCGU----------------------CGGUUGGAGGGAGGCAGGCAGUCACUGACAGUUGGA (((((.(((((((.....((.((((((.((........)----------------------).)))))).))...)))))))....)).)))..... ( -17.60) >consensus CUGCGUUUGCUUGAUUGGCUGUCCGAUAUAAGGUGUCGGACUCUUUACCUGGCCAGCAGAGCCGGAGCAGGCGGGCAAACAGGUGCCGUCAAGCGGA .....((((((((((.(((.((((((((.....)))))))).....(((((.........((........)).......)))))))))))))))))) (-25.27 = -26.61 + 1.34)


| Location | 20,892,161 – 20,892,258 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -15.83 |
| Energy contribution | -18.39 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20892161 97 - 22407834 UCCGCUUGACGGCACCUGUUUCCCCGCCUGCUCUGGCUCUGCUGGCCAGGUAAAAAGUCCGACACCUUAUAUCGGACAGCCAAUCAAGCAAACGCAG ...((((((.((((((((....((.((..((....))...)).)).))))).....((((((.........)))))).)))..))))))........ ( -33.30) >DroSec_CAF1 75599 97 - 1 UCCGCUUGACGGCACCUGUUUGCCCGCCUGCUCCGGCUCUGCUGGCCAGGUAAAGAGUCCGACACCUUAUAUCGGACAGCCAAUCAAGCAAACGCAG ...((((((.((((((((...(((.((..((....))...)).)))))))).....((((((.........)))))).)))..))))))........ ( -36.80) >DroSim_CAF1 79341 97 - 1 UCCGCUUGACGGCACCUGUUUGCCCGCCUGCUCCGGCUCUGCUGGCCAGGUAAAGAGUCCGACACCUUAUAUCGGACAGCCAAUCAAGCAAACGCAG ...((((((.((((((((...(((.((..((....))...)).)))))))).....((((((.........)))))).)))..))))))........ ( -36.80) >DroEre_CAF1 77090 92 - 1 UCCGCUCCACGGCACCUGUUUCCCCGC-----ACUGCUCUGCUGGCCAGGUAAAGAGACCGACACCUUAUAUCGGACAGCCAAUCAAGCAAACGCAG ...(((....((((((((....((.((-----(......))).)).))))).......((((.........))))...))).....)))........ ( -21.40) >DroYak_CAF1 68143 92 - 1 UCCGCUUGACGGCACCUGUUUCCCCGC----UACUGCUCUGCUG-CCAGGUAAAAAGUCCGACACCUUAUAUCGGACAGCCAAUCAAGCAAACGCAG ...((((((.((((((((.......((----....)).......-.))))).....((((((.........)))))).)))..))))))........ ( -28.66) >DroAna_CAF1 63952 75 - 1 UCCAACUGUCAGUGACUGCCUGCCUCCCUCCAACCG----------------------ACGACACCUUCUAUCGGACAACCAAUCAAGCAAACGCAG ...........(((..(((.((...........(((----------------------(.((.....))..)))).........)).)))..))).. ( -7.85) >consensus UCCGCUUGACGGCACCUGUUUCCCCGCCUGCUACGGCUCUGCUGGCCAGGUAAAGAGUCCGACACCUUAUAUCGGACAGCCAAUCAAGCAAACGCAG ...((((((.((((((((.......((........)).........))))).....((((((.........)))))).)))..))))))........ (-15.83 = -18.39 + 2.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:59 2006