
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,876,833 – 20,876,958 |
| Length | 125 |
| Max. P | 0.927729 |

| Location | 20,876,833 – 20,876,934 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.97 |
| Mean single sequence MFE | -16.64 |
| Consensus MFE | -9.89 |
| Energy contribution | -11.17 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20876833 101 - 22407834 GAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGUUUAA----UAAAACACACGGCGGGCAUAAU-AAA-GUAUCGAAU-----------AUAAAAAGCAACCACACUCAAC ((.(((...(((((((((.........(...)..(((((((....----......))))))))))))))))-...-)))))....-----------...................... ( -21.00) >DroSec_CAF1 65910 101 - 1 GAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGUUUAA----UAAAACACACGGCGGGCAUAAU-AAA-GUAUCGAAU-----------AUAAAAAGCGACCCCACUCAAC ((.(((...(((((((((.........(...)..(((((((....----......))))))))))))))))-...-)))))....-----------...................... ( -21.00) >DroSim_CAF1 69858 101 - 1 GAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGUUUAA----UAAAACACACGGCGGGCAUAAU-AAA-GUAUCGAAU-----------AUAAAAAGCGACCCCACUCAAC ((.(((...(((((((((.........(...)..(((((((....----......))))))))))))))))-...-)))))....-----------...................... ( -21.00) >DroEre_CAF1 67375 114 - 1 GAAGGCAAAAUUAUGCCCAUAAUAAAAUAAACGUGCUGUGUUUAAAGUUUAAAGCACACGGCGAGCAUAAUUAAGUAUAAGGAAUAUAUAUGGAAUAUAAAAAGCGACCC----CAAU ...((((......))))..............((((((((((..............))))))).......(((..(((((.......)))))..))).......)))....----.... ( -16.74) >DroYak_CAF1 58842 91 - 1 AAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGCUUAA----UAAAACACACGGCGGGCAAGAAUGAG-GUACCGAAUAUAUGUA----------------------UAAU ..(((((......(((((.........(...)..((((((.....----.......))))))))))).......(-....).......))))----------------------)... ( -16.00) >DroPer_CAF1 75468 86 - 1 GAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGUUGUA----G----AGAGACAGAGAGAGGGA-----------GAAGCCAU-------------AUAAAGAGAAUCGAAUCAAU ....(((......)))...........((..((((((...----.----...))))......((..-----------....))..-------------...........))..))... ( -4.10) >consensus GAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGUUUAA____UAAAACACACGGCGGGCAUAAU_AAA_GUAUCGAAU___________AUAAAAAGCGACCCCACUCAAC ..........((((((((...((.........))((((((................))))))))))))))................................................ ( -9.89 = -11.17 + 1.28)

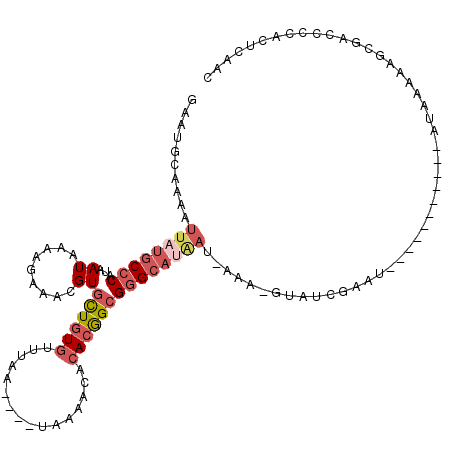
| Location | 20,876,862 – 20,876,958 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -17.15 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20876862 96 + 22407834 AC-UUU-AUUAUGCCCGCCGUGUGUUUUA----UUAAACACAGCACGUUUCUUUUAUUAUGGGCAUAAUUUUGCAUUCAAUUAAAACGGCGAGCAAAAAACC ..-...-....(((.((((((((((((..----..)))))).....................(((......)))...........)))))).)))....... ( -23.50) >DroSec_CAF1 65939 96 + 1 AC-UUU-AUUAUGCCCGCCGUGUGUUUUA----UUAAACACAGCACGUUUCUUUUAUUAUGGGCAUAAUUUUGCAUUCAAUUAAAACGGCGAGCAAAAAACC ..-...-....(((.((((((((((((..----..)))))).....................(((......)))...........)))))).)))....... ( -23.50) >DroSim_CAF1 69887 96 + 1 AC-UUU-AUUAUGCCCGCCGUGUGUUUUA----UUAAACACAGCACGUUUCUUUUAUUAUGGGCAUAAUUUUGCAUUCAAUUAAAACGGCGAGCAGAAAACC ..-...-....(((.((((((((((((..----..)))))).....................(((......)))...........)))))).)))....... ( -23.50) >DroEre_CAF1 67411 100 + 1 AUACUUAAUUAUGCUCGCCGUGUGCUUUAAACUUUAAACACAGCACGUUUAUUUUAUUAUGGGCAUAAUUUUGCCUUCGAUUA-AACGGCGAGCAAAAA-CC ...........(((((((((((((((...............))))).........(((..(((((......)))))..)))..-.))))))))))....-.. ( -31.76) >DroYak_CAF1 58860 96 + 1 AC-CUCAUUCUUGCCCGCCGUGUGUUUUA----UUAAGCACAGCACGUUUCUUUUAUUAUGGGCAUAAUUUUGCAUUUAAUUAAAACGGCGAGCAAAAA-AC ..-.......((((.((((((((((((..----..))))))..........(((.((((...(((......)))...)))).))))))))).))))...-.. ( -25.30) >DroAna_CAF1 56958 81 + 1 -----------------UUGGCUGUUGUA----UUUAGUGCAACACGUUUCGUUUAUAAUGGGCAUAAUUUUGCAUUCAAUUUAAAUGGCUAGGAAAAAACC -----------------((((((((((((----(...)))))))).....((((((...((.(((......)))...))...)))))))))))......... ( -16.40) >consensus AC_UUU_AUUAUGCCCGCCGUGUGUUUUA____UUAAACACAGCACGUUUCUUUUAUUAUGGGCAUAAUUUUGCAUUCAAUUAAAACGGCGAGCAAAAAACC ...........(((.((((((((((((........)))))).....................(((......)))...........)))))).)))....... (-17.15 = -17.35 + 0.20)



| Location | 20,876,862 – 20,876,958 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -23.41 |
| Consensus MFE | -15.84 |
| Energy contribution | -18.32 |
| Covariance contribution | 2.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20876862 96 - 22407834 GGUUUUUUGCUCGCCGUUUUAAUUGAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGUUUAA----UAAAACACACGGCGGGCAUAAU-AAA-GU .......(((((((((((((.((((...(((......)))...)))).))))........(((((((..----..))))))))))))))))....-...-.. ( -24.70) >DroSec_CAF1 65939 96 - 1 GGUUUUUUGCUCGCCGUUUUAAUUGAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGUUUAA----UAAAACACACGGCGGGCAUAAU-AAA-GU .......(((((((((((((.((((...(((......)))...)))).))))........(((((((..----..))))))))))))))))....-...-.. ( -24.70) >DroSim_CAF1 69887 96 - 1 GGUUUUCUGCUCGCCGUUUUAAUUGAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGUUUAA----UAAAACACACGGCGGGCAUAAU-AAA-GU .......(((((((((((((.((((...(((......)))...)))).))))........(((((((..----..))))))))))))))))....-...-.. ( -24.60) >DroEre_CAF1 67411 100 - 1 GG-UUUUUGCUCGCCGUU-UAAUCGAAGGCAAAAUUAUGCCCAUAAUAAAAUAAACGUGCUGUGUUUAAAGUUUAAAGCACACGGCGAGCAUAAUUAAGUAU ..-....((((((((((.-........((((......))))...............(((((...............)))))))))))))))........... ( -30.26) >DroYak_CAF1 58860 96 - 1 GU-UUUUUGCUCGCCGUUUUAAUUAAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGCUUAA----UAAAACACACGGCGGGCAAGAAUGAG-GU ..-(((((((((((((((((.((((...(((......)))...))))....)))))(((.(((......----....))))))))))))))))))....-.. ( -27.10) >DroAna_CAF1 56958 81 - 1 GGUUUUUUCCUAGCCAUUUAAAUUGAAUGCAAAAUUAUGCCCAUUAUAAACGAAACGUGUUGCACUAAA----UACAACAGCCAA----------------- ((((.((((...((.((((.....))))))....(((((.....)))))..)))).(((((......))----)))...))))..----------------- ( -9.10) >consensus GGUUUUUUGCUCGCCGUUUUAAUUGAAUGCAAAAUUAUGCCCAUAAUAAAAGAAACGUGCUGUGUUUAA____UAAAACACACGGCGGGCAUAAU_AAA_GU .......((((((((((...........(((......))).....................((((((........))))))))))))))))........... (-15.84 = -18.32 + 2.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:56 2006