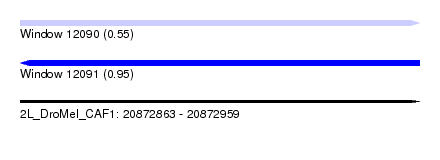
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,872,863 – 20,872,959 |
| Length | 96 |
| Max. P | 0.952248 |

| Location | 20,872,863 – 20,872,959 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -25.86 |
| Energy contribution | -27.25 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
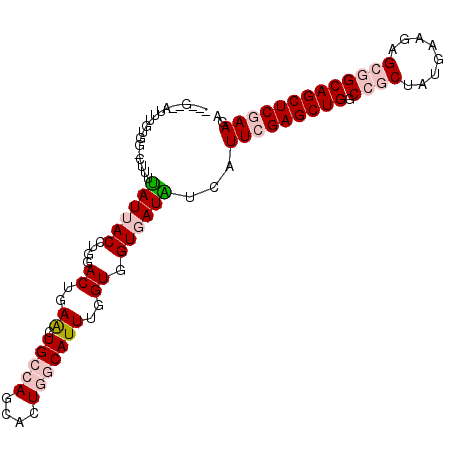
>2L_DroMel_CAF1 20872863 96 + 22407834 ------GCCCAUGG-GUUUCAUUACCUGGACUGAACUGGAAGCACUGGCAUUUGGUGGUGAUGUCGUUCGAGCUGGCCGCGAUGAAGAGCGGCAGCUCGAAGA ------(((....)-))..(((((((........((..((.((....)).))..)))))))))...(((((((((.((((........))))))))))))).. ( -37.90) >DroVir_CAF1 75648 102 + 1 GGUUCUGUUAGGAGAGU-UUAUUACCUGGACUGAACUGCCAGCACUGGCAUUUCGUUGUUAUAUCAUUCGAGCUGGCCGCUAUAAAGAGGGGCAGCUCGAAGA (((((.((((((.((..-...)).))).))).)))))((((....)))).................(((((((((.((.((......)))).))))))))).. ( -35.90) >DroPse_CAF1 65669 98 + 1 ---G--AUGUGUGGACUUUUAUUACCUGGACUGAGCUGCCAGCACUGGCAUUUGGUUGUGAUAUCAUUCGAGCUGGCAGCUAUGAAGAGCGGCAGCUCAAAGA ---.--.......((....((((((...(((..((.(((((....)))))))..)))))))))....))((((((.(.(((......)))).))))))..... ( -31.50) >DroYak_CAF1 54893 102 + 1 GGCGAUGCUCGUGG-GUUUUAUUACCUGGACUGAACUGGAAGCACUGGCAUUUGGUGGUGAUGUCGUUCGAGCUGGCCGCGAUGAAGAGCGGCAGCUCGAAGA .((((((.((..((-((......))))..(((..((..((.((....)).))..)))))))))))))((((((((.((((........))))))))))))... ( -37.80) >DroAna_CAF1 52522 96 + 1 ------AUUUGUGG-CUUUUAUUACCUGGACUGAACUGCCAGCACUGGCAUUUGGUGGUUAUGUCGUUCGAGCUGGCCGCUAUGAAGAGGGGCAGCUCGAAGA ------......((-(.......(((...((..((.(((((....)))))))..)))))...))).(((((((((.((.((......)))).))))))))).. ( -34.60) >DroPer_CAF1 69708 98 + 1 ---G--AUGUGUGGACUUUUAUUACCUGGACUGAGCUGCCAGCACUGGCAUUUGGUUGUGAUAUCAUUCGAGCUGGCAGCUAUGAAGAGCGGCAGCUCAAAGA ---.--.......((....((((((...(((..((.(((((....)))))))..)))))))))....))((((((.(.(((......)))).))))))..... ( -31.50) >consensus ___G__AUUUGUGG_CUUUUAUUACCUGGACUGAACUGCCAGCACUGGCAUUUGGUGGUGAUAUCAUUCGAGCUGGCCGCUAUGAAGAGCGGCAGCUCGAAGA ...................((((((....((..((.(((((....)))))))..)).))))))...(((((((((.((((........))))))))))))).. (-25.86 = -27.25 + 1.39)


| Location | 20,872,863 – 20,872,959 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20872863 96 - 22407834 UCUUCGAGCUGCCGCUCUUCAUCGCGGCCAGCUCGAACGACAUCACCACCAAAUGCCAGUGCUUCCAGUUCAGUCCAGGUAAUGAAAC-CCAUGGGC------ (((((((((((((((........)))).))))))))).))................................((((((((......))-)..)))))------ ( -29.90) >DroVir_CAF1 75648 102 - 1 UCUUCGAGCUGCCCCUCUUUAUAGCGGCCAGCUCGAAUGAUAUAACAACGAAAUGCCAGUGCUGGCAGUUCAGUCCAGGUAAUAA-ACUCUCCUAACAGAACC (((((((((((((.((......)).)).))))))))).)).............(((((....)))))((((.((..(((......-.....))).)).)))). ( -29.50) >DroPse_CAF1 65669 98 - 1 UCUUUGAGCUGCCGCUCUUCAUAGCUGCCAGCUCGAAUGAUAUCACAACCAAAUGCCAGUGCUGGCAGCUCAGUCCAGGUAAUAAAAGUCCACACAU--C--- ((((((((((((.(((......))).).))))))))).)).............((((.(..((((....))))..).))))................--.--- ( -24.30) >DroYak_CAF1 54893 102 - 1 UCUUCGAGCUGCCGCUCUUCAUCGCGGCCAGCUCGAACGACAUCACCACCAAAUGCCAGUGCUUCCAGUUCAGUCCAGGUAAUAAAAC-CCACGAGCAUCGCC ..(((((((((((((........)))).)))))))))(((..............((....)).....((((.((...(((......))-).)))))).))).. ( -29.00) >DroAna_CAF1 52522 96 - 1 UCUUCGAGCUGCCCCUCUUCAUAGCGGCCAGCUCGAACGACAUAACCACCAAAUGCCAGUGCUGGCAGUUCAGUCCAGGUAAUAAAAG-CCACAAAU------ ..(((((((((((.((......)).)).))))))))).(((..(((.......(((((....))))))))..)))..(((.......)-))......------ ( -27.21) >DroPer_CAF1 69708 98 - 1 UCUUUGAGCUGCCGCUCUUCAUAGCUGCCAGCUCGAAUGAUAUCACAACCAAAUGCCAGUGCUGGCAGCUCAGUCCAGGUAAUAAAAGUCCACACAU--C--- ((((((((((((.(((......))).).))))))))).)).............((((.(..((((....))))..).))))................--.--- ( -24.30) >consensus UCUUCGAGCUGCCGCUCUUCAUAGCGGCCAGCUCGAACGACAUCACAACCAAAUGCCAGUGCUGGCAGUUCAGUCCAGGUAAUAAAAC_CCACAAAC__C___ (((((((((((((((........)))).))))))))).)).............((((.(..(((......)))..).))))...................... (-25.04 = -25.82 + 0.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:53 2006