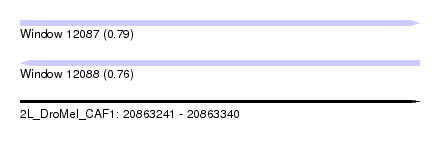
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,863,241 – 20,863,340 |
| Length | 99 |
| Max. P | 0.786089 |

| Location | 20,863,241 – 20,863,340 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
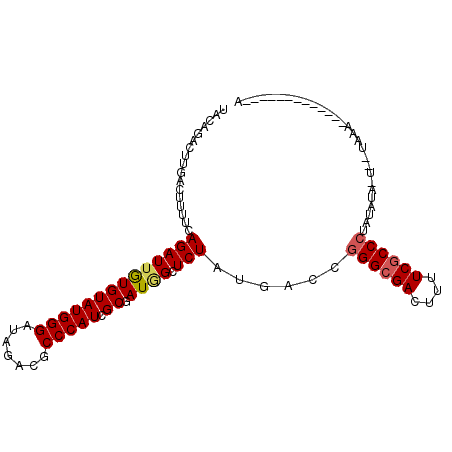
>2L_DroMel_CAF1 20863241 99 + 22407834 UACAAGCUUGACUUUUCAGAUUGUGUAUGGGAUAAACGCCCAUCGCGAUUGCUCUAUGACCGGGCGAUUUUUCACCCUACAGU-UUAUACAUAAUCAUG-----A ...............((((((((((((((((((((.(((.....))).))).)))......(((.((....)).)))......-.))))))))))).))-----) ( -23.90) >DroVir_CAF1 59864 88 + 1 UACAGACUUGACUUUUCAGAUUGUGUAUGGGAUAGACACCCAUGGCGAUGGCUCUAUGACCGGGCGACUUUUCGCCCUGUAUACA----CA-------------A .......((((....)))).((((((((((.(((((...((((....)))).)))))..))((((((....))))))...)))))----))-------------) ( -27.00) >DroPse_CAF1 50823 90 + 1 UACAGACUUGACUUUUCAGAUUGUGUAUGGGAUAGACGCCCAUCGCGAUAGCUCUAUGACCGGGCGACUUUUCGCCCUAUAUA-UGGUAA--------------A .......((((....))))((((((.(((((.......))))))))))).(((.((((...((((((....))))))..))))-.)))..--------------. ( -25.40) >DroGri_CAF1 41136 88 + 1 UACAGACUUGACUUUUGAGAUUGUGUAUGGGAUAGACGCCCAUGGCGAUGGCUCUAUGACCGGGCGACUUUUCGCCCUGUUUAGU----GG-------------U .....(((.(((....((((((((.((((((.......)))))))))))..))).......((((((....)))))).))).)))----..-------------. ( -25.80) >DroWil_CAF1 64293 103 + 1 CACAAACUUUACUUUUCAGAUUAUGUAUGGGAUAAACGCCCAUGGCAAUGGCUCUAUGACCAGGCGACUUUUCGCCCUAUAU--AUAUAUAUAGUAUUUGGUCAA .................((((((((((((((.......))))).)).)))).))).(((((((((((....)))))((((((--....))))))....)))))). ( -26.80) >DroPer_CAF1 53672 90 + 1 UACAGACUUGACUUUUCAGAUUGUGUAUGGGAUAGACGCCCAUCGCGAUAGCUCUAUGACCGGGCGACUUUUCGCCCUACAUA-UGGUAA--------------A .......((((....))))((((((.(((((.......))))))))))).(((.((((...((((((....))))))..))))-.)))..--------------. ( -27.20) >consensus UACAGACUUGACUUUUCAGAUUGUGUAUGGGAUAGACGCCCAUCGCGAUGGCUCUAUGACCGGGCGACUUUUCGCCCUAUAUA_U__UAAA_____________A .................((((((((((((((.......))))).)).)))).)))......((((((....))))))............................ (-16.99 = -17.18 + 0.20)

| Location | 20,863,241 – 20,863,340 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
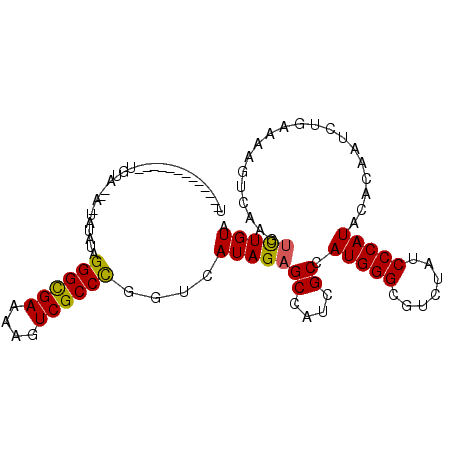
>2L_DroMel_CAF1 20863241 99 - 22407834 U-----CAUGAUUAUGUAUAA-ACUGUAGGGUGAAAAAUCGCCCGGUCAUAGAGCAAUCGCGAUGGGCGUUUAUCCCAUACACAAUCUGAAAAGUCAAGCUUGUA (-----((.((((.(((((((-((....((((((....))))))(.((((.((....))...)))).)))))).....)))).))))))).(((.....)))... ( -24.10) >DroVir_CAF1 59864 88 - 1 U-------------UG----UGUAUACAGGGCGAAAAGUCGCCCGGUCAUAGAGCCAUCGCCAUGGGUGUCUAUCCCAUACACAAUCUGAAAAGUCAAGUCUGUA (-------------((----(((((...((((((....))))))((..(((((..((((......))))))))).)))))))))).((....))........... ( -26.80) >DroPse_CAF1 50823 90 - 1 U--------------UUACCA-UAUAUAGGGCGAAAAGUCGCCCGGUCAUAGAGCUAUCGCGAUGGGCGUCUAUCCCAUACACAAUCUGAAAAGUCAAGUCUGUA .--------------......-......((((((....))))))....(((((((....)).(((((.......)))))....................))))). ( -20.80) >DroGri_CAF1 41136 88 - 1 A-------------CC----ACUAAACAGGGCGAAAAGUCGCCCGGUCAUAGAGCCAUCGCCAUGGGCGUCUAUCCCAUACACAAUCUCAAAAGUCAAGUCUGUA .-------------..----(((..((.((((((....))))))((..((((((((.........))).))))).))................))..)))..... ( -22.30) >DroWil_CAF1 64293 103 - 1 UUGACCAAAUACUAUAUAUAU--AUAUAGGGCGAAAAGUCGCCUGGUCAUAGAGCCAUUGCCAUGGGCGUUUAUCCCAUACAUAAUCUGAAAAGUAAAGUUUGUG .((((((....((((((....--))))))(((((....)))))))))))((((.....((..(((((.......))))).))...))))................ ( -26.40) >DroPer_CAF1 53672 90 - 1 U--------------UUACCA-UAUGUAGGGCGAAAAGUCGCCCGGUCAUAGAGCUAUCGCGAUGGGCGUCUAUCCCAUACACAAUCUGAAAAGUCAAGUCUGUA .--------------......-((((..((((((....))))))...))))..((....)).(((((.......)))))..(((..((((....)).))..))). ( -21.50) >consensus U_____________UGUA__A_UAUAUAGGGCGAAAAGUCGCCCGGUCAUAGAGCCAUCGCCAUGGGCGUCUAUCCCAUACACAAUCUGAAAAGUCAAGUCUGUA ............................((((((....))))))....(((((((....)).(((((.......)))))....................))))). (-18.55 = -18.22 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:51 2006