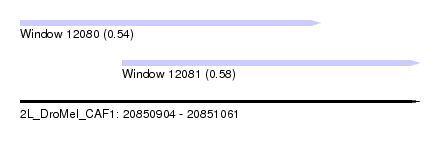
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,850,904 – 20,851,061 |
| Length | 157 |
| Max. P | 0.584394 |

| Location | 20,850,904 – 20,851,022 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -24.86 |
| Energy contribution | -24.87 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20850904 118 + 22407834 AGAUUUUUACUCUUUGCCUAAAUUUAACCCGUUGUAUAUCAUAUGCUCAACUUGCUUGUCAAGUGAGUCAAGCUACAUCGGUAUGCAGAGGGAUCCCCAUUACCUGUCGGCCUCCAGA-- ...............(((.........(((.(((((((((....(((..(((..((.....))..)))..)))......))))))))).))).....((.....))..))).......-- ( -26.50) >DroSec_CAF1 29424 120 + 1 AGAUUUUUACUCCUUGCCCAAUAUCAAGCCUUUGUAUACCACAUGCUCAACUCGCUUGUCAAGUAAGUCAAGUUACAUCGGUAUGCAGAGGGAUCCCCAUUACCUGUAGACCUCCAGACU .((..(((((..((((........))))((((((((((((....(((..(((..((.....))..)))..)))......))))))))))))..............)))))..))...... ( -28.00) >DroSim_CAF1 30581 118 + 1 AGAUUUUUACUCCUUGCCCAAUAUCAAGCCUUUGUAUACCACAUGCUCAACUCGCUUGCCAAGUAAGUCAAGUUACAUCGGUAUGCAGAGGGAUCCCCAUUACCUGUAGACCUCCAGG-- .((((.......((((........))))((((((((((((....(((..(((..((.....))..)))..)))......)))))))))))))))).......((((........))))-- ( -29.40) >consensus AGAUUUUUACUCCUUGCCCAAUAUCAAGCCUUUGUAUACCACAUGCUCAACUCGCUUGUCAAGUAAGUCAAGUUACAUCGGUAUGCAGAGGGAUCCCCAUUACCUGUAGACCUCCAGA__ .((((.......((((........))))((((((((((((....(((..(((..((.....))..)))..)))......))))))))))))))))........(((........)))... (-24.86 = -24.87 + 0.01)

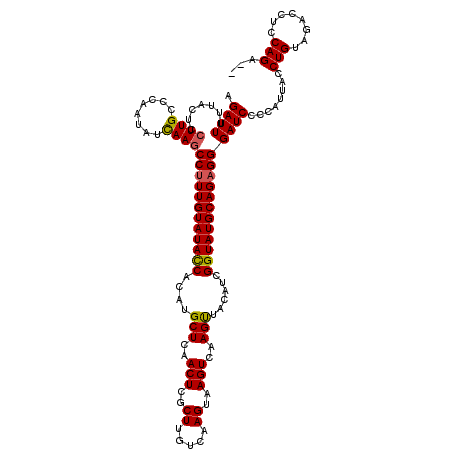
| Location | 20,850,944 – 20,851,061 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -32.41 |
| Energy contribution | -32.30 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20850944 117 + 22407834 AUAUGCUCAACUUGCUUGUCAAGUGAGUCAAGCUACAUCGGUAUGCAGAGGGAUCCCCAUUACCUGUCGGCCUCCAGA---UAGAGUCCGUUAAUGUUCUUGGCCGACUCGGGAUCCAUU .....(((.(((..((.....))..)))...(((((....))).)).)))(((((((.......((((........))---))(((((.(((((.....))))).))))))))))))... ( -37.10) >DroSec_CAF1 29464 120 + 1 ACAUGCUCAACUCGCUUGUCAAGUAAGUCAAGUUACAUCGGUAUGCAGAGGGAUCCCCAUUACCUGUAGACCUCCAGACUCCAGAGUCCGUUACUGUUCUUGGCGGACUCGGGAUCCAUU .((((((......(((((.(......).)))))......)))))).....(((((((......(((........)))......((((((((((.......)))))))))))))))))... ( -40.00) >DroSim_CAF1 30621 117 + 1 ACAUGCUCAACUCGCUUGCCAAGUAAGUCAAGUUACAUCGGUAUGCAGAGGGAUCCCCAUUACCUGUAGACCUCCAGG---UAGAGUCCGUUACUGUUCUUGGCCGCCUCGGGAUCCAUU ..........(((((.((((..((((......))))...)))).)).)))(((((((...((((((........))))---))(((.(.((((.......)))).).))))))))))... ( -35.70) >consensus ACAUGCUCAACUCGCUUGUCAAGUAAGUCAAGUUACAUCGGUAUGCAGAGGGAUCCCCAUUACCUGUAGACCUCCAGA___UAGAGUCCGUUACUGUUCUUGGCCGACUCGGGAUCCAUU .((((((......(((((.(......).)))))......)))))).....(((((((......(((........)))......(((((.((((.......)))).))))))))))))... (-32.41 = -32.30 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:45 2006