
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
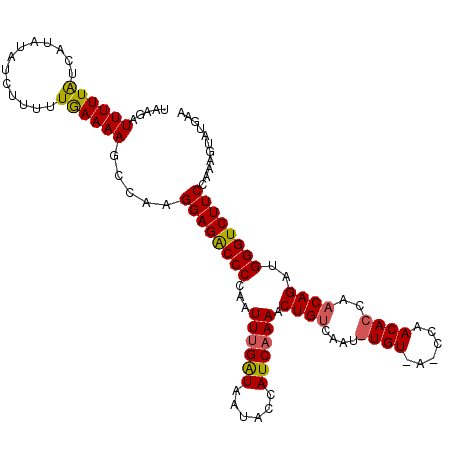
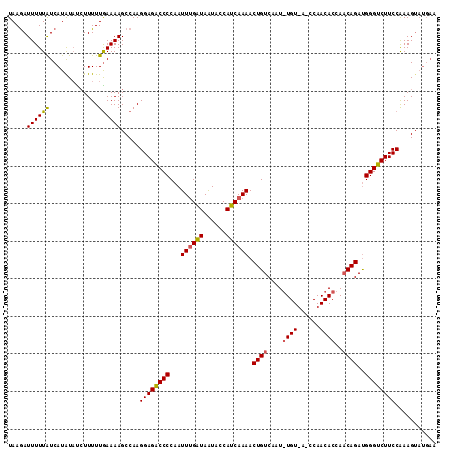
| Location | 20,845,723 – 20,845,831 |
| Length | 108 |
| Max. P | 0.977345 |

| Location | 20,845,723 – 20,845,831 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -15.83 |
| Energy contribution | -16.29 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20845723 108 + 22407834 UAAGAUUUUUAUCAUAUAUCUUUUUGAAAAGCCAAGGAGGCCCCAAUUCGAUAAUACCAUCAAAACUGUCAAUGUGUAAGCCAACACCACCAGACGGGUCUUCCAAAG------ ...(((....)))......................(((((.(((.....(((......)))......(((...((((......)))).....)))))).)))))....------ ( -19.50) >DroSec_CAF1 23841 114 + 1 UAAAAUUUUUGUCAUAUAUCUUUUUAAAAAGCCAAGGAGACCCCAAUUUGAUAAUACCAUCAAAACUGUCAAUAUGUUAUCCAACACCAACAGAUGGGUCUUCCGAAGUAUGAA ...........((((((..((((....))))....((((((((...((((((......)))))).((((.....((((....))))...))))..))))))))....)))))). ( -23.20) >DroSim_CAF1 24727 96 + 1 UAAGAUUUUUAUCAUGUUCAUUUUUGAAAAGCCAAGGAGACCCCAAUUUGGUAGUACCAUCAAAACUGUCA------------------ACAGAUGGGUCUUCCAAAGUAUGAA ...........(((((((((....)))).......((((((((...((((((......)))))).(((...------------------.)))..)))))))).....))))). ( -22.00) >consensus UAAGAUUUUUAUCAUAUAUCUUUUUGAAAAGCCAAGGAGACCCCAAUUUGAUAAUACCAUCAAAACUGUCAAU_UGU_A_CCAACACCAACAGAUGGGUCUUCCAAAGUAUGAA .....((((((.............)))))).....((((((((...((((((......)))))).((((....((((......))))..))))..))))))))........... (-15.83 = -16.29 + 0.45)

| Location | 20,845,723 – 20,845,831 |
|---|---|
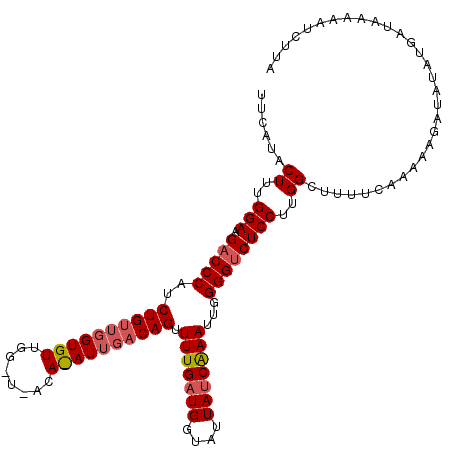
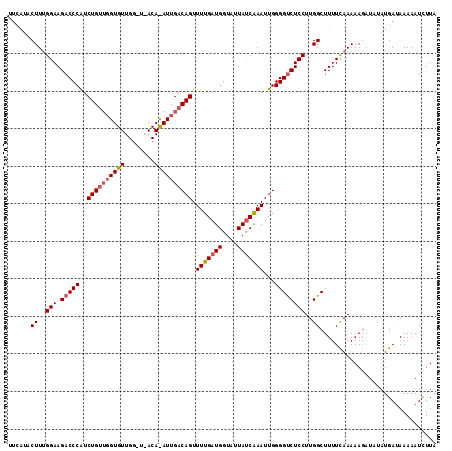
| Length | 108 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -18.10 |
| Energy contribution | -19.77 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20845723 108 - 22407834 ------CUUUGGAAGACCCGUCUGGUGGUGUUGGCUUACACAUUGACAGUUUUGAUGGUAUUAUCGAAUUGGGGCCUCCUUGGCUUUUCAAAAAGAUAUAUGAUAAAAAUCUUA ------.((((((((.((((((.....((((......))))...)))...(((((((....)))))))..)))(((.....)))))))))))(((((...........))))). ( -24.90) >DroSec_CAF1 23841 114 - 1 UUCAUACUUCGGAAGACCCAUCUGUUGGUGUUGGAUAACAUAUUGACAGUUUUGAUGGUAUUAUCAAAUUGGGGUCUCCUUGGCUUUUUAAAAAGAUAUAUGACAAAAAUUUUA .(((((((..(((.(((((..((((..((((.(.....)))))..)))).(((((((....)))))))...))))))))..))((((....))))...)))))........... ( -28.80) >DroSim_CAF1 24727 96 - 1 UUCAUACUUUGGAAGACCCAUCUGU------------------UGACAGUUUUGAUGGUACUACCAAAUUGGGGUCUCCUUGGCUUUUCAAAAAUGAACAUGAUAAAAAUCUUA .((((.((..(((.(((((..(((.------------------...)))......(((.....))).....))))))))..))...((((....)))).))))........... ( -20.60) >consensus UUCAUACUUUGGAAGACCCAUCUGUUGGUGUUGG_U_ACA_AUUGACAGUUUUGAUGGUAUUAUCAAAUUGGGGUCUCCUUGGCUUUUCAAAAAGAUAUAUGAUAAAAAUCUUA ......((..(((.(((((..((((((((((........)))))))))).(((((((....)))))))...))))))))..))............................... (-18.10 = -19.77 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:42 2006