
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,831,938 – 20,832,036 |
| Length | 98 |
| Max. P | 0.597501 |

| Location | 20,831,938 – 20,832,036 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -16.86 |
| Energy contribution | -16.70 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20831938 98 - 22407834 ---U--CUCU--GGCCAAUUUAAACGUCAAACGUUAACGUUCGGGUUAAAUGCACUUAGCCAACACUU-UGA-GGUGCGUUUAAACGAAAGCCU----AGAUAAGUCGAA---------A ---.--.(((--(((.......((((.....))))....((((..((((((((((((...(((....)-)))-))))))))))).)))).))).----))).........---------. ( -24.50) >DroVir_CAF1 11103 107 - 1 ---U--GUGG--GGCCAAUUUAAACGUCAAACGUUAACGUUCAGGUUAAAUGCACUUAACCAACACUU-UGA-GGUGCGUUUAAAUGAAAGCUU----AGAUAAACAGCAAAUAGCCUGG ---.--...(--(((..((((((((((...((.((((.((...((((((......))))))...)).)-)))-.))))))))))))....(((.----........))).....)))).. ( -24.80) >DroGri_CAF1 9871 107 - 1 ---U--GCGG--GGCCAAUUUAAACGUCAAACGUUAACGUUCAGGUUAAAUGCACUUAACCAACACUU-UGA-GGUGCGUUUAAAUGAAAGCUC----AGAUAAGCAGCAAACAUCAACA ---(--((.(--(((..((((((((((...((.((((.((...((((((......))))))...)).)-)))-.))))))))))))....))))----......)))............. ( -25.10) >DroWil_CAF1 9873 115 - 1 GUUUGCCUCA--AGUCAAUUUAAACGUCAAGCGUUAACGUUCAGGUUAAAUGCACUUAACCAACACUUUUGA-GGUGCGUUUAAAUGAAAGCCAUGAAAGAUAAGCAAAA--UCCCCAUG .(((((....--..(((.....((((.....))))....((((..((((((((((((...(((.....))))-))))))))))).)))).....))).......))))).--........ ( -23.12) >DroMoj_CAF1 11722 109 - 1 ---U--GUGCUUUGCCAAUUUAAACGUCAAACGUUAACGUUCAGGUUAAAUGCACUUAACUAACACUU-UGA-GGUGCGUUUAAAUGAAAGCUU----AGAUAAACAGCAAAUGGCCAAG ---.--...(((.((((.....((((.....))))....((((..((((((((((((...........-..)-))))))))))).)))).(((.----........)))...)))).))) ( -24.92) >DroAna_CAF1 9170 99 - 1 ---U--CGCC--GGCCAAUUUAAACGUCAAGCUCUAACGUUCGGGUUAAAUGCACUUAGCCAACACUU-UGAAGGUGCGUUUAAAUGAAAGCCU----AGAUAAGUCGGA---------G ---.--..((--(((..(((((....(((.((((........))))(((((((((((...(((....)-)).)))))))))))..))).....)----))))..))))).---------. ( -23.50) >consensus ___U__CUCC__GGCCAAUUUAAACGUCAAACGUUAACGUUCAGGUUAAAUGCACUUAACCAACACUU_UGA_GGUGCGUUUAAAUGAAAGCCU____AGAUAAGCAGCA__U__CCA_G ............(((..(((((((((...((((....))))..((((((......))))))..((((......)))))))))))))....)))........................... (-16.86 = -16.70 + -0.16)

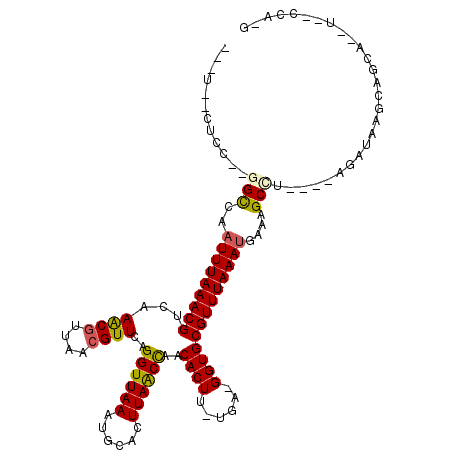
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:39 2006