
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,825,248 – 20,825,488 |
| Length | 240 |
| Max. P | 0.948232 |

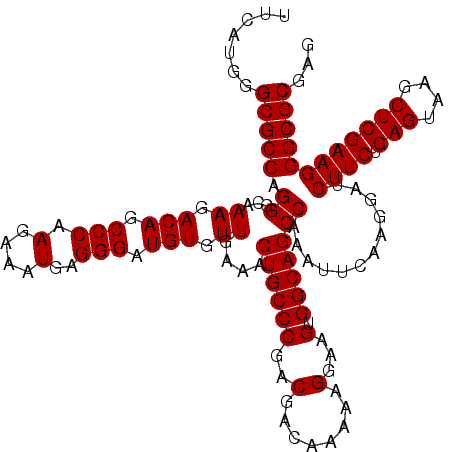
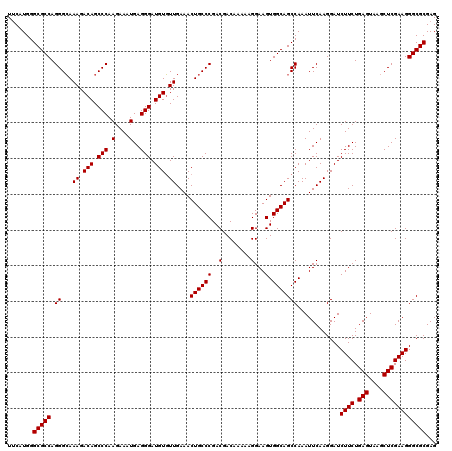
| Location | 20,825,248 – 20,825,368 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -34.20 |
| Energy contribution | -34.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20825248 120 - 22407834 UACAUGGGCGCCAGGGGAAAGACAGCCCAAGAAAUGAGGGAUGUUUUGAAACUGCCCGACGACAAGAAGGAAGUGGCAGCCAAAUUCAAGGAUCUUCUGAGUAAACUCGAAGGGCGCGAG .......(((((.((...((((((.(((.(....)..))).))))))....((((((..(........)...).)))))))............((((.(((....))))))))))))... ( -36.20) >DroSec_CAF1 3788 120 - 1 UUCAUGGGCGCCAGGGCAAAGACAGCCCAAGAAAUGAGGGAUGUGUUGAAACUGCCCGACGACAAAAAGGAAGUGGCAGCCAAAUUCAAGGAUCUUCUGAGUAAGCUCGAAGGGCGCGAG .......(((((.((((.......))))......((((...((.((((..(((..((...........)).)))..))))))..)))).....((((.(((....))))))))))))... ( -36.90) >DroSim_CAF1 3804 120 - 1 UUCAUGGGCGCCAGGGCAAAGACAGCCCAAGAAAUGAGGGAUGUGUUGAAACUGCCCGACGACAAAAAGGAAGUGGCAGCCAAAUUCAAGGAUCUUCUGAGUAAGCUCGAAGGGCGCGAG .......(((((.((((.......))))......((((...((.((((..(((..((...........)).)))..))))))..)))).....((((.(((....))))))))))))... ( -36.90) >consensus UUCAUGGGCGCCAGGGCAAAGACAGCCCAAGAAAUGAGGGAUGUGUUGAAACUGCCCGACGACAAAAAGGAAGUGGCAGCCAAAUUCAAGGAUCUUCUGAGUAAGCUCGAAGGGCGCGAG .......(((((.((...((.(((.(((.(....)..))).))).))....((((((..(........)...).)))))))............((((.(((....))))))))))))... (-34.20 = -34.20 + -0.00)

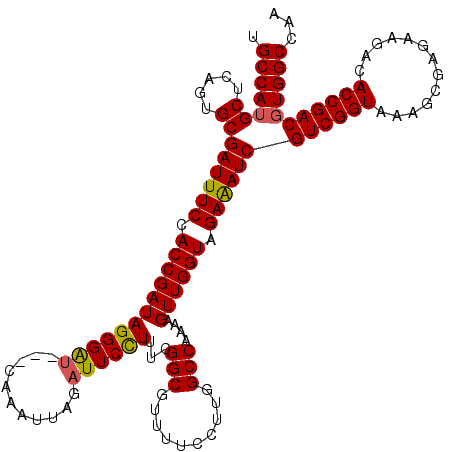
| Location | 20,825,368 – 20,825,488 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -43.53 |
| Consensus MFE | -34.55 |
| Energy contribution | -34.55 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20825368 120 + 22407834 UGCCAGGCUCAGUGCGAUUUCCACCGAUAGGGGAGAGGUAAUGAGAUUCUUGUCGGCAUUUUCCUUGGCCAAAAGUUGGUAGAGAUCGUCGGUGAAGCGACAAGACACCGACGUGGCCAA .((((.((.....))((((((.((((((..((..((((.((((.(((....)))..))))..))))..))....)))))).))))))(((((((...(.....).))))))).))))... ( -43.90) >DroSec_CAF1 3908 117 + 1 UGCCAUGCUCAGUGCGAUUUCCACCGAUAGGGAU---CAAAUUAGAUUCCUUUCGGCGUUUUCCUUGGCCAAAAGUUGGUAGAAAUCGUCGGUAAAGCGAGAAGUCACCGACGUGGCCAA .(((((((.....))((((((.(((((.((((((---(......))))))).))(((..........))).......))).))))))((((((...((.....)).)))))))))))... ( -43.20) >DroSim_CAF1 3924 116 + 1 UGCCAUGCUCAGUGCGAUUUCCACCGAUAGGGAU---CAAAUUAGAUUCCUUUCGGCGUUUUCCUGGGCCAAACGUUGGUAGAAAUCGUCGGUAAAGCGAGAAGACACCGACGUGGCCA- .(((((((.....))((((((.(((((.((((((---(......))))))).))(((.(......).))).......))).))))))((((((....(.....)..)))))))))))..- ( -43.50) >consensus UGCCAUGCUCAGUGCGAUUUCCACCGAUAGGGAU___CAAAUUAGAUUCCUUUCGGCGUUUUCCUUGGCCAAAAGUUGGUAGAAAUCGUCGGUAAAGCGAGAAGACACCGACGUGGCCAA .(((((((.....))((((((.((((((((((((...........))))))...(((..........)))....)))))).))))))((((((.............)))))))))))... (-34.55 = -34.55 + 0.01)


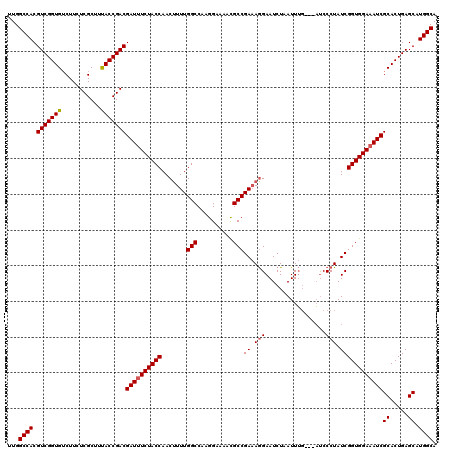
| Location | 20,825,368 – 20,825,488 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -40.37 |
| Consensus MFE | -35.12 |
| Energy contribution | -35.56 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20825368 120 - 22407834 UUGGCCACGUCGGUGUCUUGUCGCUUCACCGACGAUCUCUACCAACUUUUGGCCAAGGAAAAUGCCGACAAGAAUCUCAUUACCUCUCCCCUAUCGGUGGAAAUCGCACUGAGCCUGGCA ...((((.((((((((...((((......))))(((.((((((.....(((((..........)))))...(.....).................)))))).)))))))))).).)))). ( -34.30) >DroSec_CAF1 3908 117 - 1 UUGGCCACGUCGGUGACUUCUCGCUUUACCGACGAUUUCUACCAACUUUUGGCCAAGGAAAACGCCGAAAGGAAUCUAAUUUG---AUCCCUAUCGGUGGAAAUCGCACUGAGCAUGGCA ...((((.((((((((.........))))))))((((((((((....((((((..........))))))(((.(((......)---)).)))...))))))))))((.....)).)))). ( -43.50) >DroSim_CAF1 3924 116 - 1 -UGGCCACGUCGGUGUCUUCUCGCUUUACCGACGAUUUCUACCAACGUUUGGCCCAGGAAAACGCCGAAAGGAAUCUAAUUUG---AUCCCUAUCGGUGGAAAUCGCACUGAGCAUGGCA -..((((.(((((((...........)))))))((((((((((..(((((..(....).)))))..((.(((.(((......)---)).))).))))))))))))((.....)).)))). ( -43.30) >consensus UUGGCCACGUCGGUGUCUUCUCGCUUUACCGACGAUUUCUACCAACUUUUGGCCAAGGAAAACGCCGAAAGGAAUCUAAUUUG___AUCCCUAUCGGUGGAAAUCGCACUGAGCAUGGCA ...((((.(((((((...........)))))))((((((((((.......(((..........)))((.(((.................))).))))))))))))((.....)).)))). (-35.12 = -35.56 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:38 2006