| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,149,466 – 2,149,613 |
| Length | 147 |
| Max. P | 0.832144 |

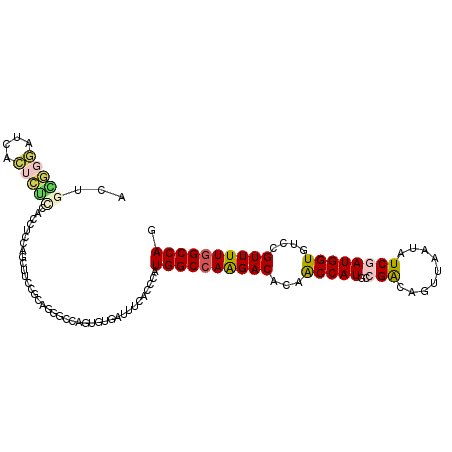
| Location | 2,149,466 – 2,149,583 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -25.59 |
| Energy contribution | -26.48 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2149466 117 - 22407834 CGUAGGGUCAGUGUUAUUUCUCCCAUGGCUAAGACACAACCAUGCCGGCAGUUAAUAUCGAUGGUGUCAGUUUUUGCCAGCAGUUGCGGCAGCACCACAGCGGCCAUGUACUCGGGC .((((((..((......))..)))((((((............(((.(((((...((((.....))))......))))).)))((((.((.....)).)))))))))).)))...... ( -29.70) >DroVir_CAF1 3454 117 - 1 CGCAGCGCCAGGGUUAUCUCGCCCAUGGCCAAAACACAGCCAUGACGAGCGUUCACAUCAAUCGUUUCGGUCUUGGCCAAGAGCUGGGGCAUAACCACAGCGGCCAUGUACUCUGGC ......((((((((.........(((((((........((((.((((((((...........)))))..))).)))).....((((((......)).))))))))))).)))))))) ( -45.20) >DroWil_CAF1 3411 117 - 1 CGCAGAGCCAAAGUAAUUUCACCCAUGGCCAAGACACAACCAUGACGAGAAUUAACAUCUAUGGUGUCGGUUUUGGCCAAUAAUUGUGGUAGCACCACCGCGGCCAUAUACUCUGGC ......((((.((((..........((((((((((...(((((((.(........).)).)))))....)))))))))).....((((((.((......)).)))))))))).)))) ( -40.30) >DroMoj_CAF1 3490 117 - 1 CUUAGGGCCAGCGUUAUCUCGCCCAUGGCCAGCACGCAGCCAUGCCUUGCGUUCACGUCGAUCGUCUCCGUCUUGGCCAAGAGUUGAGGCAGGACAACGGCCGCCAUGUACUCGGGU ....((((.((......)).))))..((((.(((.(((....)))..)))(((((((.....)))....((((..((.....))..)))).))))...))))(((.((....))))) ( -37.30) >DroAna_CAF1 4706 117 - 1 CUCAGGGCCAGGGUGAUCUCUCCCAUGGCCAGGACACAACCAUGUCGGCAGUUGAUGUCAAUGGUAUCCGUUUUGGCCAAGAGCUGCGGCAACACCACUGCCGCCAUAUAUUCCGGC (((.(((..((((....))))))).((((((((((...(((((...((((.....)))).)))))....)))))))))).)))..((((((.......))))))............. ( -43.00) >DroPer_CAF1 3391 117 - 1 CGGACGGCCAGGGUGAUUUCACCCAUGGCCAGGACACAACCAUGCCGACUGUUGAUGUCGAUGGUGUCCGUUUUGGCCAGGAGUUGAGGCAGCACAACGGCCGCCAUAUACUCUGGC (((((((((((((((....))))).)))))........(((((..((((.......)))))))))))))).....(((((.(((...(((.((......)).)))....)))))))) ( -53.60) >consensus CGCAGGGCCAGGGUUAUCUCACCCAUGGCCAAGACACAACCAUGCCGACAGUUAACAUCGAUGGUGUCCGUUUUGGCCAAGAGUUGAGGCAGCACCACAGCCGCCAUAUACUCUGGC ......(((.((((...........((((((((((...(((((.................)))))....))))))))))........(((.((......)).)))....)))).))) (-25.59 = -26.48 + 0.89)

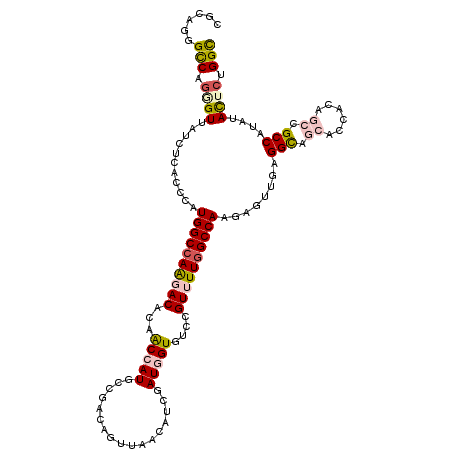
| Location | 2,149,503 – 2,149,613 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2149503 110 - 22407834 ACUUGGGGAUCACUCUUUUCCUCUAGUUUCCGUAGGGUCAGUGUUAUUUCUCCCAUGGCUAAGACACAACCAUGCCGGCAGUUAAUAUCGAUGGUGUCAGUUUUUGCCAG ...((((((.((((....(((((........).))))..)))).....)).))))((((.(((((...(((((..((..(.....)..)))))))....))))).)))). ( -26.50) >DroPse_CAF1 3425 110 - 1 ACGGCUGGAUCACUGGCCAUUUCCAGCUUGCGGACGGCCAGUGUGAUUUCACCCAUGGCCAGGACACAACCAUGCCGACUGUUGAUGUCGAUGGUGUCCGUUUUGGCCAG ..((.((((.((((((((...(((.......))).))))))))....)))).)).((((((((((...(((((..((((.......)))))))))....)))))))))). ( -48.50) >DroEre_CAF1 3625 110 - 1 ACUUGGGGAUCACUCUUUUCCUCCAGCUUGCGCAGGGCCAGUGUUAUUUCUCCCAUGGCCAAGACACAGCCAUGCCGGCAGUUAAUAUCGAUGGUGUCAGUUUUGGCCAG ...((((((.((((....((((...((....))))))..)))).....)).))))((((((((((...(((((..((..(.....)..)))))))....)))))))))). ( -33.70) >DroWil_CAF1 3448 110 - 1 GUUGCACCUUUAGAGUCCAGCUCCAGUUGCCGCAGAGCCAAAGUAAUUUCACCCAUGGCCAAGACACAACCAUGACGAGAAUUAACAUCUAUGGUGUCGGUUUUGGCCAA ((.(((.((...(((.....))).)).))).))......................((((((((((...(((((((.(........).)).)))))....)))))))))). ( -29.70) >DroAna_CAF1 4743 110 - 1 ACAGAGUUCUCACUCUUCUGCUCCAAAUUCCUCAGGGCCAGGGUGAUCUCUCCCAUGGCCAGGACACAACCAUGUCGGCAGUUGAUGUCAAUGGUAUCCGUUUUGGCCAA ..((((...(((((((...((((...........)))).))))))).))))....((((((((((...(((((...((((.....)))).)))))....)))))))))). ( -38.20) >DroPer_CAF1 3428 110 - 1 AUGGCUGGAUCACUGGCCAUUUCCAGCUUGCGGACGGCCAGGGUGAUUUCACCCAUGGCCAGGACACAACCAUGCCGACUGUUGAUGUCGAUGGUGUCCGUUUUGGCCAG .((((..((...((((......))))...((((((((((((((((....))))).)))))........(((((..((((.......)))))))))))))))))..)))). ( -51.90) >consensus ACUGCGGGAUCACUCUCCACCUCCAGCUUCCGCAGGGCCAGUGUGAUUUCACCCAUGGCCAAGACACAACCAUGCCGACAGUUAAUAUCGAUGGUGUCCGUUUUGGCCAG ...(((((....)))))......................................((((((((((...(((((..(((.........))))))))....)))))))))). (-20.33 = -20.33 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:45 2006