
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,809,534 – 20,809,694 |
| Length | 160 |
| Max. P | 0.877785 |

| Location | 20,809,534 – 20,809,654 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -29.54 |
| Energy contribution | -29.38 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20809534 120 + 22407834 UGCCUCAAUGUUGCGCGUCAAUUUUAGGGAGCCUACACCAUAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCCCACUUUUUUUUCAUCUUGGCCUUCUCGACAGGC .((((....((((.(.((((((((((.((........)).)))))...((.(((((((((((((.......)))).))))).))))))..............)))))...).)))))))) ( -34.10) >DroSec_CAF1 32552 120 + 1 UGCCUCAAUGUUGCGCGUCAAUUUUAGGGAGCCUACACUAUAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCACACUUUCCUUUCAUCUUGGCCUCCAUGGCUGUC .(((...(((.....)))........((..(((...........((...(((((((((((((((.......)))).))))).))))))...))...........)))..))..))).... ( -35.65) >DroSim_CAF1 36926 120 + 1 UGCCUCAAUGUUGCGCGUCAAUUUUAGGGAGCCUACACUAUAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCCCACUUUCCUUUCAUCUUGGCCUCCUUGGCCGGA .(((........).))..........(((((((...........(((((....)))))((((((.......)))))).)))...))))..............((((((.....)))))). ( -38.20) >DroEre_CAF1 35069 120 + 1 UGCCUCAAUGUUGCGCGUCAAUUUUAGGGAGCCUACACUAGAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCUCGUCGCACUUUCCUUUCUUCUUGGCCUCCUUGGCUGCC ............(((.((((.....(((..(((......(((((.....(((((((((((((((.......)))).))))).)))))).......)))))....)))..)))))))))). ( -40.20) >DroYak_CAF1 33516 120 + 1 UGCCUCAAUGUUGCGCGUCAAUUUUAGGGAGCCUACACUAUAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCCCACUUUCCGUUCUUCUUGGCCAUCUUGGCUGGC .(((......................(((((((...........(((((....)))))((((((.......)))))).)))...))))................((((.....))))))) ( -37.20) >consensus UGCCUCAAUGUUGCGCGUCAAUUUUAGGGAGCCUACACUAUAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCCCACUUUCCUUUCAUCUUGGCCUCCUUGGCUGGC .(((........).))((((.....(((..(((...(((.........)))(((((((((((((.......)))).))))).))))..................)))..))))))).... (-29.54 = -29.38 + -0.16)

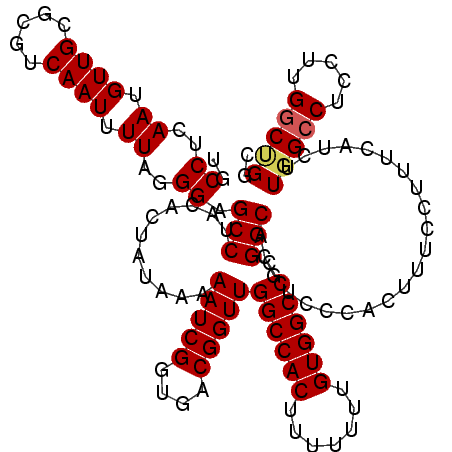

| Location | 20,809,534 – 20,809,654 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -30.30 |
| Energy contribution | -31.22 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20809534 120 - 22407834 GCCUGUCGAGAAGGCCAAGAUGAAAAAAAAGUGGGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUAUGGUGUAGGCUCCCUAAAAUUGACGCGCAACAUUGAGGCA (((((((((...((((.(.((.(.(((((..(((((((.(...((((((.......))))))..))))).))).))))).).)).).))))........)))))...((....)))))). ( -36.30) >DroSec_CAF1 32552 120 - 1 GACAGCCAUGGAGGCCAAGAUGAAAGGAAAGUGUGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUAUAGUGUAGGCUCCCUAAAAUUGACGCGCAACAUUGAGGCA ....(((..((((.(((..(((((((....(.((((((.(...((((((.......))))))..))))))))..)))))))..))..).))))......(((.....)))......))). ( -37.70) >DroSim_CAF1 36926 120 - 1 UCCGGCCAAGGAGGCCAAGAUGAAAGGAAAGUGGGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUAUAGUGUAGGCUCCCUAAAAUUGACGCGCAACAUUGAGGCA ....(((.(((..(((.(.((.....((((((((((((.(...((((((.......))))))..))))).)).))))))...)).).)))..)))....(((.....)))......))). ( -38.50) >DroEre_CAF1 35069 120 - 1 GGCAGCCAAGGAGGCCAAGAAGAAAGGAAAGUGCGACGAGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUCUAGUGUAGGCUCCCUAAAAUUGACGCGCAACAUUGAGGCA ....(((.(((..(((.(.(....(((((((((.((((.(...((((((.......))))))..))))).)..))))))))..).).)))..)))....(((.....)))......))). ( -36.10) >DroYak_CAF1 33516 120 - 1 GCCAGCCAAGAUGGCCAAGAAGAACGGAAAGUGGGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUAUAGUGUAGGCUCCCUAAAAUUGACGCGCAACAUUGAGGCA (((.(((.....)))...........((((((((((((.(...((((((.......))))))..))))).)).)))))).((((((.(((((.........)).)).).)))))).))). ( -36.00) >consensus GCCAGCCAAGGAGGCCAAGAUGAAAGGAAAGUGGGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUAUAGUGUAGGCUCCCUAAAAUUGACGCGCAACAUUGAGGCA ....(((.(((.((((.(.((....(((((.(((((((.(...((((((.......))))))..))))).))).)))))...)).).)))).)))....(((.....)))......))). (-30.30 = -31.22 + 0.92)

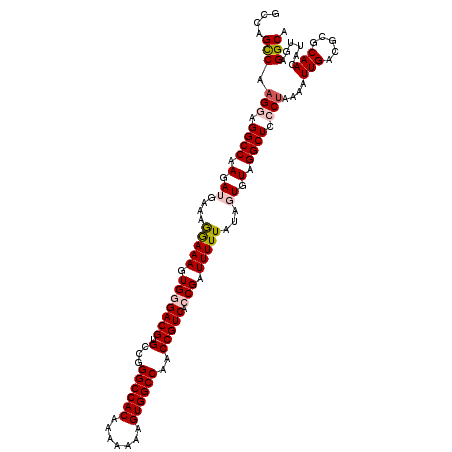
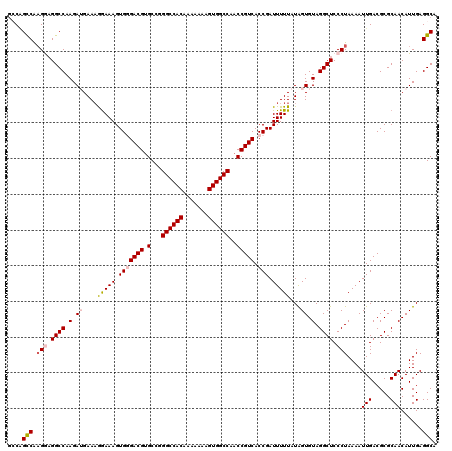
| Location | 20,809,574 – 20,809,694 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -49.86 |
| Consensus MFE | -37.68 |
| Energy contribution | -37.32 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20809574 120 + 22407834 UAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCCCACUUUUUUUUCAUCUUGGCCUUCUCGACAGGCCGUUGAGUGGAGUUGGGCGCUCCAAAAGCGCCUGCGCGGA ....(((((....)))))((((((.......)))))).((..(((((.(((((...((((...((((((.......)))))).)))).))))).)))))..))....(((....)))... ( -48.60) >DroSec_CAF1 32592 120 + 1 UAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCACACUUUCCUUUCAUCUUGGCCUCCAUGGCUGUCCGCUGGGUGGAGUUGGGCACUCCGAAGGCGCCUGCGCGGA .........(((((((((((((((.......)))).))))).))))))................((((.....)))).(((((((((((...((((.....))))...)))))).))))) ( -47.60) >DroSim_CAF1 36966 120 + 1 UAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCCCACUUUCCUUUCAUCUUGGCCUCCUUGGCCGGACGCUGGGUGGAGUUGGACACUCCGAAGGCGCCUGCGCGGA ....(((((....)))))((((((.......))))))(((...((((.((((((((..(.(((.((((.....))))))).)..))).))))).))))...)))...(((....)))... ( -49.40) >DroEre_CAF1 35109 120 + 1 GAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCUCGUCGCACUUUCCUUUCUUCUUGGCCUCCUUGGCUGCCUGCGGAGUGGAGUUGGGCGCUCCGAAGGCGCCUGCGCGGA .........(((((((((((((((.......)))).))))).))))))....(((.........((((.....))))((((.(((((((........))))))).))))((....))))) ( -50.60) >DroYak_CAF1 33556 120 + 1 UAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCCCACUUUCCGUUCUUCUUGGCCAUCUUGGCUGGCCGCAGAGUGGAGUUGGGCGCUCGGAAGGCGCCUGCGCGGA ........((.(((((((((((((.......)))).))))).))))))....(((((((((((((((((.......)))))).)))..)))...((((((.(....)))))))..))))) ( -53.10) >consensus UAAAAAUCGGUGACGGUUGGCCACUUUUUUUGUGGCCCGGCACGUCCCACUUUCCUUUCAUCUUGGCCUCCUUGGCUGGCCGCUGAGUGGAGUUGGGCGCUCCGAAGGCGCCUGCGCGGA ......((((((((((((((((((.......)))).))))).)))).....((((.((((.(.(((((.....)))))...).)))).))))..(((((((.....))))))))).))). (-37.68 = -37.32 + -0.36)

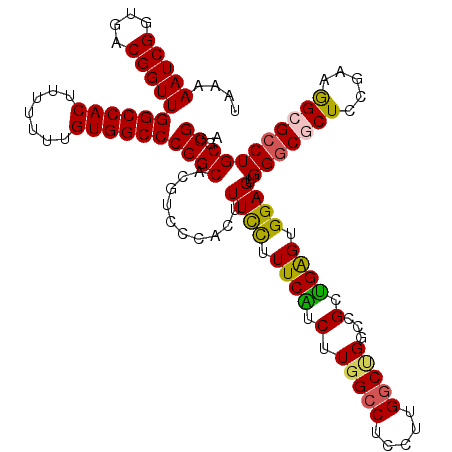
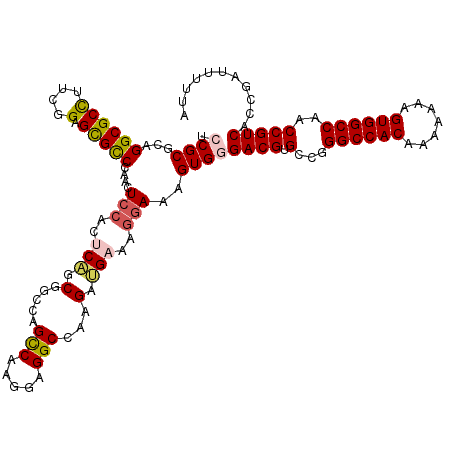
| Location | 20,809,574 – 20,809,694 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -33.44 |
| Energy contribution | -33.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20809574 120 - 22407834 UCCGCGCAGGCGCUUUUGGAGCGCCCAACUCCACUCAACGGCCUGUCGAGAAGGCCAAGAUGAAAAAAAAGUGGGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUA .((((...(((((((...)))))))..............(((((.......)))))..............))))((((.(...((((((.......))))))..)))))........... ( -43.10) >DroSec_CAF1 32592 120 - 1 UCCGCGCAGGCGCCUUCGGAGUGCCCAACUCCACCCAGCGGACAGCCAUGGAGGCCAAGAUGAAAGGAAAGUGUGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUA (((...((.(.(((((((((((.....))))).....((.....))...)))))))....))...)))..(.((((((.(...((((((.......))))))..))))))))........ ( -44.40) >DroSim_CAF1 36966 120 - 1 UCCGCGCAGGCGCCUUCGGAGUGUCCAACUCCACCCAGCGUCCGGCCAAGGAGGCCAAGAUGAAAGGAAAGUGGGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUA (((...(((((((....(((((.....))))).....))))).((((.....))))....))...)))...(((((((.(...((((((.......))))))..))))).)))....... ( -45.80) >DroEre_CAF1 35109 120 - 1 UCCGCGCAGGCGCCUUCGGAGCGCCCAACUCCACUCCGCAGGCAGCCAAGGAGGCCAAGAAGAAAGGAAAGUGCGACGAGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUC ...((((.((((((....).)))))....(((..((....(((..(....)..))).....))..)))..))))((((.(...((((((.......))))))..)))))........... ( -44.40) >DroYak_CAF1 33556 120 - 1 UCCGCGCAGGCGCCUUCCGAGCGCCCAACUCCACUCUGCGGCCAGCCAAGAUGGCCAAGAAGAACGGAAAGUGGGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUA ((((..(.((((((....).))))).........(((..(((((.......))))).))).)..))))...(((((((.(...((((((.......))))))..))))).)))....... ( -45.30) >consensus UCCGCGCAGGCGCCUUCGGAGCGCCCAACUCCACUCAGCGGCCAGCCAAGGAGGCCAAGAUGAAAGGAAAGUGGGACGUGCCGGGCCACAAAAAAAGUGGCCAACCGUCACCGAUUUUUA .((((...((((((....).)))))....(((..(((.(.....(((.....)))...).)))..)))..))))((((.(...((((((.......))))))..)))))........... (-33.44 = -33.88 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:30 2006