| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,803,583 – 20,803,739 |
| Length | 156 |
| Max. P | 0.777866 |

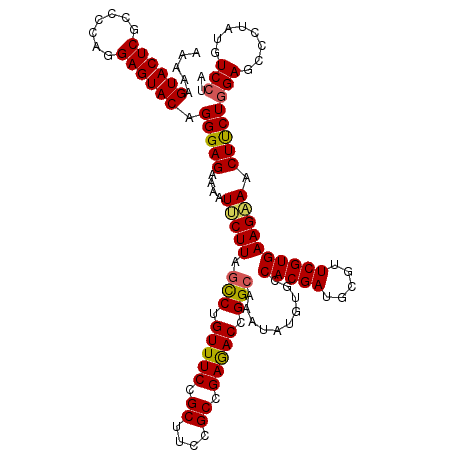
| Location | 20,803,583 – 20,803,703 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -32.64 |
| Energy contribution | -32.52 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20803583 120 - 22407834 AAAAAGUACUCGCCCCAGGAGUACAGGGAGAAAAUUCUUAGCCUGUUUCCGCUUCCGCCGAGACCGGCAAAUAUGUGCCACGAUGCGUUCGUGAAGAAACUUCUGGAGCCCUAUGUCCUA .....((((((.......))))))((((((((..(((((.(((.(((((.((....)).))))).)))..........(((((.....))))))))))..))))....))))........ ( -37.50) >DroSec_CAF1 26845 120 - 1 AAAAAGUACUCGCCCCAGGAGUACAGGGAGAAAAUUCUUAGCCUGUUUCCGCUUCCGCCGAGACCGGCACAUAUGUGCCACGAUGCGUUCGUGAAGAAACUUCUGGAGCCCUAUGUCCUA .....((((((.......))))))((((((((..(((((.(((.(((((.((....)).))))).)))..........(((((.....))))))))))..))))....))))........ ( -37.50) >DroSim_CAF1 30428 120 - 1 AAAAAGUACUCGCCCCAGGAGUACAGGGAGAAAAUUCUUAGCCUGUUUCCGCUUCCGCCGAGACCGGCACAUAUGUGCCACGAUGCGUUCGUGAAGAAACUUCUGGAGCCCUAUGUCCUA .....((((((.......))))))((((((((..(((((.(((.(((((.((....)).))))).)))..........(((((.....))))))))))..))))....))))........ ( -37.50) >DroEre_CAF1 30243 120 - 1 AAGAAGUACUCACCACAGGAGUACAGGGAGAAAAUCCUUAGCCUGUUUCCGCUUCCGCCGAAACCGGAAAAUAUGUGCCACGAUGCGUUCGUGAAGGAGCUCCUGGAGCCAUAUGUCCUG .....((((((.......)))))).(((((....(((((..((.(((((.((....)).))))).))...........(((((.....)))))))))).)))))(((........))).. ( -37.80) >DroYak_CAF1 27741 120 - 1 AAAAAGUACUCACCUCAGGAGUACAGGGAGAAGAUCCUUAGUCUGUUUCCGCUUCCGCCGAGACCGGCAAAUAUGUGCCACGAUGCGUUCGUGAAGAAACUUCUCGAGCCAUAUGUCCUA .....((((((.......)))))).((((.(((((.....))))(((((.((....)).))))).((((......))))...(((.(((((.((((...)))).)))))))).).)))). ( -35.50) >consensus AAAAAGUACUCGCCCCAGGAGUACAGGGAGAAAAUUCUUAGCCUGUUUCCGCUUCCGCCGAGACCGGCAAAUAUGUGCCACGAUGCGUUCGUGAAGAAACUUCUGGAGCCCUAUGUCCUA .....((((((.......)))))).(((((....(((((.(((.(((((.((....)).))))).)))..........(((((.....)))))))))).)))))(((........))).. (-32.64 = -32.52 + -0.12)

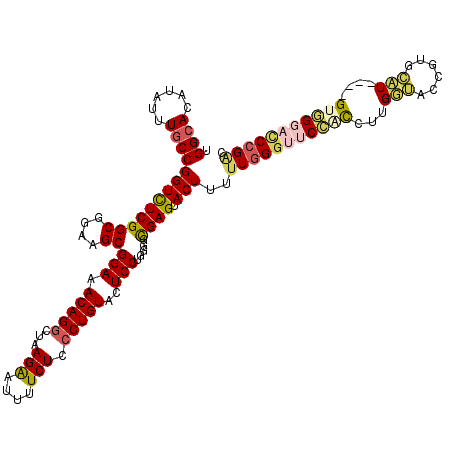
| Location | 20,803,623 – 20,803,739 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -44.36 |
| Consensus MFE | -32.36 |
| Energy contribution | -33.16 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20803623 116 + 22407834 UGGCACAUAUUUGCCGGUCUCGGCGGAAGCGGAAACAGGCUAAGAAUUUUCUCCCUGUACUCCUGGGGCGAGUACUUUUUGGGUUCCACCUUGAUACCGUGCAU----GUGGGACCCGCU .(((...((((((((.......((....))(((.(((((...(((....))).)))))..)))...))))))))......(((((((((..((........)).----)))))))))))) ( -43.00) >DroSec_CAF1 26885 116 + 1 UGGCACAUAUGUGCCGGUCUCGGCGGAAGCGGAAACAGGCUAAGAAUUUUCUCCCUGUACUCCUGGGGCGAGUACUUUUUGGGUUCCGCCUUGGUACCGUGACU----GUGGGACCCGAC .(((((....)))))(((((((.(((..((((..(((((...(((....))).)))))....(..(((((.(.(((.....))).))))))..)..))))..))----)))))))).... ( -48.50) >DroSim_CAF1 30468 116 + 1 UGGCACAUAUGUGCCGGUCUCGGCGGAAGCGGAAACAGGCUAAGAAUUUUCUCCCUGUACUCCUGGGGCGAGUACUUUUUGGGUUCCGCCUUGGUACCGUGACU----GUGGGACCCGAC .(((((....)))))(((((((.(((..((((..(((((...(((....))).)))))....(..(((((.(.(((.....))).))))))..)..))))..))----)))))))).... ( -48.50) >DroEre_CAF1 30283 120 + 1 UGGCACAUAUUUUCCGGUUUCGGCGGAAGCGGAAACAGGCUAAGGAUUUUCUCCCUGUACUCCUGUGGUGAGUACUUCUUGGGCUCCACCUUGUGGGAGUUCAUUUCAGGAGUUUCAGGC .............((.(((((.((....)).))))).))..............((((.(((((((..((((..((((((.(((.....)))...))))))))))..)))))))..)))). ( -44.00) >DroYak_CAF1 27781 116 + 1 UGGCACAUAUUUGCCGGUCUCGGCGGAAGCGGAAACAGACUAAGGAUCUUCUCCCUGUACUCCUGAGGUGAGUACUUUUUGGGUUCGAUCUUGUGAUUGUGCAU----GGCGCUACCGAG ..(((((...(..(.(((((..((....))(....)))))).((((((....(((.(((((((....).)))))).....)))...)))))))..).))))).(----((.....))).. ( -37.80) >consensus UGGCACAUAUUUGCCGGUCUCGGCGGAAGCGGAAACAGGCUAAGAAUUUUCUCCCUGUACUCCUGGGGCGAGUACUUUUUGGGUUCCACCUUGGUACCGUGCAU____GUGGGACCCGAC .((((......))))(((((((((....))(((.(((((...(((....))).)))))..))).....)))).)))..(((((((((((...(((......)))....))))))))))). (-32.36 = -33.16 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:24 2006