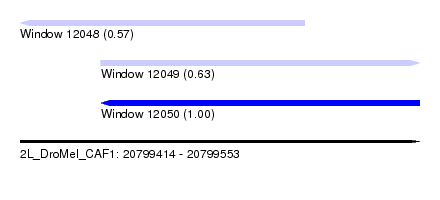
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,799,414 – 20,799,553 |
| Length | 139 |
| Max. P | 0.996865 |

| Location | 20,799,414 – 20,799,513 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -18.52 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20799414 99 - 22407834 UGAUUCAAAGGAAAAACAUUCAAGAUUUGUACAGC-------CGCGGACAAAUCUCAUG--UUUGCCGCCUCUAAUGGCCAUGAAAA------------CCUGUGACCAUGGUGGUCAAC ...((((..((..((((((...((((((((.(...-------...).)))))))).)))--))).))(((......)))..))))..------------....(((((.....))))).. ( -25.60) >DroSec_CAF1 22691 106 - 1 UGAUUCAAAGGAAAAACAUUUAAGAUUUGUACAGCAAAGCGUCGCAGACAAAUCUCAUG--UUUGCCGCCUCUAAUGGCCAUGAAAA------------CACUGGUCUUCGGUGGUCAAC (((((((..((..((((((...((((((((...((........))..)))))))).)))--))).))(((......)))..)))...------------(((((.....))))))))).. ( -27.10) >DroSim_CAF1 26277 106 - 1 UGAUUCAAAGGAAAAACAUUUAAGAUUUGUACAGCAAAGCGUCGCGGACAAAUCUCAUG--UUUGCCGCCUCUAAUGGCCAUGAAAA------------CACUGGUCUUCGGUGGUCAAC (((((((..((..((((((...((((((((...((........))..)))))))).)))--))).))(((......)))..)))...------------(((((.....))))))))).. ( -28.20) >DroEre_CAF1 26195 106 - 1 UGAUUCAAAGGAAAAACAUUCAAGAUUUGUACAGCAAAGCGCCGCGAACAAAUCUCUUG--UUUGCCGCUUCCAAUGGCCAUGGAAA------------CCCUGGUCUUCAGCGGUCAAC .............(((((....((((((((...((........))..))))))))..))--)))((((((......(((((.((...------------.)))))))...)))))).... ( -28.50) >DroYak_CAF1 23691 120 - 1 UGCUUCAAAGGAAAAACAUUCAAGAUUUGUACGGCAAGUUGCCGCAAACAAAUCUCUUUUUUUUGCCGCCUCCAAUGGCCAUGAAAAGUGUUUUUACUUCAGUGGUCUUCAGUGGUCAAC .((..(((((((((........((((((((.((((.....))))...)))))))).)))))))))..))..(((..((((((...(((((....)))))..)))))).....)))..... ( -31.30) >consensus UGAUUCAAAGGAAAAACAUUCAAGAUUUGUACAGCAAAGCGCCGCGGACAAAUCUCAUG__UUUGCCGCCUCUAAUGGCCAUGAAAA____________CACUGGUCUUCGGUGGUCAAC ......................((((((((...((........))..)))))))).......((((((((......(((((.....................)))))...))))).))). (-18.52 = -18.92 + 0.40)

| Location | 20,799,442 – 20,799,553 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -23.21 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20799442 111 + 22407834 GGCCAUUAGAGGCGGCAAA--CAUGAGAUUUGUCCGCG-------GCUGUACAAAUCUUGAAUGUUUUUCCUUUGAAUCAGCCAUAAUUUAGUUGAAUUGUCUGAAAUGGCUGAUGAAAU .....(((((((....(((--((((((((((((.((..-------..)).)))))))))..))))))..)))))))(((((((((..(((((.........))))))))))))))..... ( -32.50) >DroSec_CAF1 22719 118 + 1 GGCCAUUAGAGGCGGCAAA--CAUGAGAUUUGUCUGCGACGCUUUGCUGUACAAAUCUUAAAUGUUUUUCCUUUGAAUCAGCCAUAAUUUAGUUGAAUUGUCUGAAAUGACUGAUGAAAU .....(((((((....(((--(((((((((((((.((((....)))).).)))))))))..))))))..)))))))(((((.(((..(((((.........)))))))).)))))..... ( -29.40) >DroSim_CAF1 26305 118 + 1 GGCCAUUAGAGGCGGCAAA--CAUGAGAUUUGUCCGCGACGCUUUGCUGUACAAAUCUUAAAUGUUUUUCCUUUGAAUCAGCCAUAAUUUAGUUGAAUUGUCUGAAAUGACUGAUGAAAU .....(((((((....(((--(((((((((((((.((((....)))).).)))))))))..))))))..)))))))(((((.(((..(((((.........)))))))).)))))..... ( -29.40) >DroEre_CAF1 26223 118 + 1 GGCCAUUGGAAGCGGCAAA--CAAGAGAUUUGUUCGCGGCGCUUUGCUGUACAAAUCUUGAAUGUUUUUCCUUUGAAUCAGCCAUAAUUUAGUUGAAUUGUCUGAGACGACUGAUGAAAU (((.(((.((((.((.(((--((.(((((((((..(((((.....))))))))))))))...))))).)))))).)))..))).....(((((((..((....))..)))))))...... ( -33.70) >DroYak_CAF1 23731 120 + 1 GGCCAUUGGAGGCGGCAAAAAAAAGAGAUUUGUUUGCGGCAACUUGCCGUACAAAUCUUGAAUGUUUUUCCUUUGAAGCAGCCAUAAUUUAGUUGAAUUGUCUGAAAUGACUGAUGAAAU ((.((((..(((((((........(((((((((..(((((.....))))))))))))))...((((((......)))))))))..(((((....)))))))))..)))).))........ ( -34.50) >consensus GGCCAUUAGAGGCGGCAAA__CAUGAGAUUUGUCCGCGACGCUUUGCUGUACAAAUCUUGAAUGUUUUUCCUUUGAAUCAGCCAUAAUUUAGUUGAAUUGUCUGAAAUGACUGAUGAAAU ...((((((....(((........((((((((((.((((....)))).).)))))))))...((.(((......))).)))))(((((((....))))))).........)))))).... (-23.21 = -22.82 + -0.39)

| Location | 20,799,442 – 20,799,553 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -23.14 |
| Energy contribution | -22.86 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
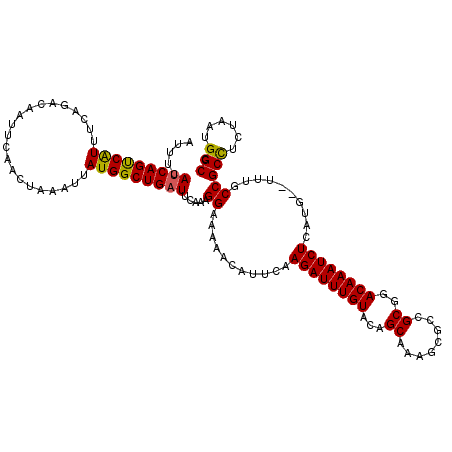
>2L_DroMel_CAF1 20799442 111 - 22407834 AUUUCAUCAGCCAUUUCAGACAAUUCAACUAAAUUAUGGCUGAUUCAAAGGAAAAACAUUCAAGAUUUGUACAGC-------CGCGGACAAAUCUCAUG--UUUGCCGCCUCUAAUGGCC .........((((((..((..................(((((...((((.(((.....)))....))))..))))-------)((((.(((((.....)--))))))))))..)))))). ( -29.40) >DroSec_CAF1 22719 118 - 1 AUUUCAUCAGUCAUUUCAGACAAUUCAACUAAAUUAUGGCUGAUUCAAAGGAAAAACAUUUAAGAUUUGUACAGCAAAGCGUCGCAGACAAAUCUCAUG--UUUGCCGCCUCUAAUGGCC .....(((((((((.....................))))))))).....((..((((((...((((((((...((........))..)))))))).)))--))).))(((......))). ( -27.60) >DroSim_CAF1 26305 118 - 1 AUUUCAUCAGUCAUUUCAGACAAUUCAACUAAAUUAUGGCUGAUUCAAAGGAAAAACAUUUAAGAUUUGUACAGCAAAGCGUCGCGGACAAAUCUCAUG--UUUGCCGCCUCUAAUGGCC .....(((((((((.....................))))))))).....((..((((((...((((((((...((........))..)))))))).)))--))).))(((......))). ( -28.70) >DroEre_CAF1 26223 118 - 1 AUUUCAUCAGUCGUCUCAGACAAUUCAACUAAAUUAUGGCUGAUUCAAAGGAAAAACAUUCAAGAUUUGUACAGCAAAGCGCCGCGAACAAAUCUCUUG--UUUGCCGCUUCCAAUGGCC .....((((((((((...)))((((......))))..))))))).....((((..........(.((((.....)))).)((.((((((((.....)))--))))).))))))....... ( -25.00) >DroYak_CAF1 23731 120 - 1 AUUUCAUCAGUCAUUUCAGACAAUUCAACUAAAUUAUGGCUGCUUCAAAGGAAAAACAUUCAAGAUUUGUACGGCAAGUUGCCGCAAACAAAUCUCUUUUUUUUGCCGCCUCCAAUGGCC .........(((......)))................(((......(((((((.....))).((((((((.((((.....))))...)))))))))))).....)))(((......))). ( -26.20) >consensus AUUUCAUCAGUCAUUUCAGACAAUUCAACUAAAUUAUGGCUGAUUCAAAGGAAAAACAUUCAAGAUUUGUACAGCAAAGCGCCGCGGACAAAUCUCAUG__UUUGCCGCCUCUAAUGGCC .....(((((((((.....................))))))))).....((...........((((((((...((........))..))))))))..........))(((......))). (-23.14 = -22.86 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:16 2006