
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,797,296 – 20,797,441 |
| Length | 145 |
| Max. P | 0.948752 |

| Location | 20,797,296 – 20,797,411 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.24 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -29.30 |
| Energy contribution | -30.38 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20797296 115 + 22407834 GCUUAGUUCGUGGCUGAAAUGCAUAUCUGGGUCAGCUGUUGUUUGGCUGUUUGUUCGACUUGCAUAAGCCAGUCCGGCGGCAUGAAAAACCCCCCAUUUCAGCCCCAAACCCAAA----- .....(((.(.((((((((((.......(((((((((((((.((((((...(((.......)))..))))))..))))))).)))....)))..)))))))))).).))).....----- ( -39.10) >DroSec_CAF1 20587 115 + 1 GUUUAGUUCGUGGCUGAAAUGCAUAUCUGGGUCAGCUGUUGUUUGGCCGUUUGUUCGACUUGCAUAAGCCAGUCCGGCGGCAUGGAAAACCCCCCAUUUCAGCCCCAAACCCAAA----- .....(((.(.((((((((((.......(((((((((((((.(((((....(((.......)))...)))))..))))))).))....))))..)))))))))).).))).....----- ( -36.00) >DroSim_CAF1 24143 109 + 1 GUUUAGUUCGUGGCUGAAAUGCAUAUCUGGGUCAGCUGUUGUUUGGCCGUUUGUUCGACUUGCAUAAGCCAGUCCGGCGGCAUGGAAAACCCCCCAUUUCAGCCCCA------AA----- .........(.((((((((((.......(((((((((((((.(((((....(((.......)))...)))))..))))))).))....))))..)))))))))).).------..----- ( -35.00) >DroEre_CAF1 24000 112 + 1 GUUCAGUUCGUGGCUGAAAUGCAUAUCUGGGUCAGCUGUUGUUUGGCCGUUUGUUCGGCUUGCAUAAGCCAGUCCGGCGGCAUG-AAAACCCCCCAUUUCAGCCCCAAACCCA------- .....(((.(.((((((((((.......(((((((((..(((..(((((......))))).)))...(((.....)))))).))-)...)))..)))))))))).).)))...------- ( -37.00) >DroYak_CAF1 21545 105 + 1 ---CGCUUCGUAGCUAAAAUACAUAUCUGGGUCAGCUGUUGUUUG------------GCUUGCAUAAGCCAGUCCGGCGGCAUGGAAAACCCCCCAUUUUAGCCCCAAACCCAAACCCCC ---.(((....))).............(((((..(((((((.(((------------((((....)))))))..)))))))((((........))))...........)))))....... ( -28.80) >consensus GUUUAGUUCGUGGCUGAAAUGCAUAUCUGGGUCAGCUGUUGUUUGGCCGUUUGUUCGACUUGCAUAAGCCAGUCCGGCGGCAUGGAAAACCCCCCAUUUCAGCCCCAAACCCAAA_____ .........(.((((((((((.......(((((((((((((.(((((....(((.......)))...)))))..))))))).))....))))..)))))))))).).............. (-29.30 = -30.38 + 1.08)


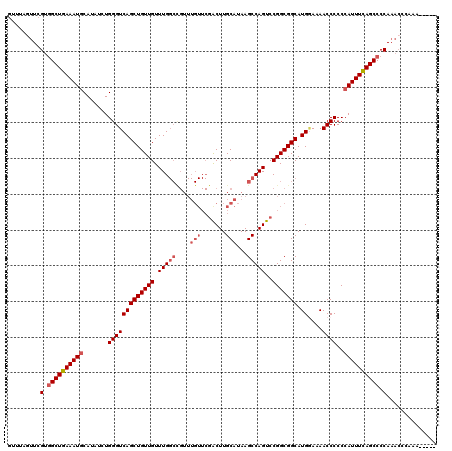
| Location | 20,797,296 – 20,797,411 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.24 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -33.54 |
| Energy contribution | -34.36 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20797296 115 - 22407834 -----UUUGGGUUUGGGGCUGAAAUGGGGGGUUUUUCAUGCCGCCGGACUGGCUUAUGCAAGUCGAACAAACAGCCAAACAACAGCUGACCCAGAUAUGCAUUUCAGCCACGAACUAAGC -----....((((((.((((((((((((.(((.......))).)).((((.((....)).)))).......((((.........))))...........)))))))))).)))))).... ( -41.20) >DroSec_CAF1 20587 115 - 1 -----UUUGGGUUUGGGGCUGAAAUGGGGGGUUUUCCAUGCCGCCGGACUGGCUUAUGCAAGUCGAACAAACGGCCAAACAACAGCUGACCCAGAUAUGCAUUUCAGCCACGAACUAAAC -----....((((((.((((((((((((.(((.......))).)).((((.((....)).)))).......((((.........))))...........)))))))))).)))))).... ( -40.50) >DroSim_CAF1 24143 109 - 1 -----UU------UGGGGCUGAAAUGGGGGGUUUUCCAUGCCGCCGGACUGGCUUAUGCAAGUCGAACAAACGGCCAAACAACAGCUGACCCAGAUAUGCAUUUCAGCCACGAACUAAAC -----((------((.((((((((((((.(((.......))).)).((((.((....)).)))).......((((.........))))...........)))))))))).))))...... ( -35.60) >DroEre_CAF1 24000 112 - 1 -------UGGGUUUGGGGCUGAAAUGGGGGGUUUU-CAUGCCGCCGGACUGGCUUAUGCAAGCCGAACAAACGGCCAAACAACAGCUGACCCAGAUAUGCAUUUCAGCCACGAACUGAAC -------(.((((((.((((((((((((.(((...-...))).))((..(((((..((...((((......))))....))..)))))..)).......)))))))))).)))))).).. ( -43.00) >DroYak_CAF1 21545 105 - 1 GGGGGUUUGGGUUUGGGGCUAAAAUGGGGGGUUUUCCAUGCCGCCGGACUGGCUUAUGCAAGC------------CAAACAACAGCUGACCCAGAUAUGUAUUUUAGCUACGAAGCG--- ..(((((.(((((((((((....((((((....))))))))).)))))))(((((....))))------------).........).)))))..............(((....))).--- ( -32.40) >consensus _____UUUGGGUUUGGGGCUGAAAUGGGGGGUUUUCCAUGCCGCCGGACUGGCUUAUGCAAGUCGAACAAACGGCCAAACAACAGCUGACCCAGAUAUGCAUUUCAGCCACGAACUAAAC .........((((((.((((((((((((.(((.......))).))((..(((((..((...((((......))))....))..)))))..)).......)))))))))).)))))).... (-33.54 = -34.36 + 0.82)


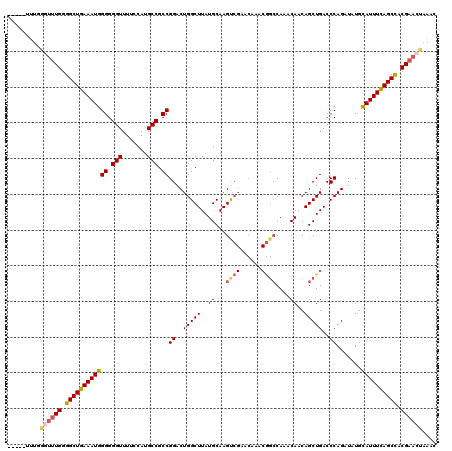
| Location | 20,797,336 – 20,797,441 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -15.60 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20797336 105 + 22407834 GUUUGGCUGUUUGUUCGACUUGCAUAAGCCAGUCCGGCGGCAUGAAAAACCCCCCAUUUCAGCCCCAAACCCAAA---------------CUCAAACCCAACCCCGUUUUUCAACCCUGC ((((((..((((....((((.((....)).)))).((.(((.(((((.........)))))))))))))))))))---------------)............................. ( -23.70) >DroSec_CAF1 20627 105 + 1 GUUUGGCCGUUUGUUCGACUUGCAUAAGCCAGUCCGGCGGCAUGGAAAACCCCCCAUUUCAGCCCCAAACCCAAA---------------CUCAAACCCAACCCCGUUUUUCAACCCUGC ((((((..(((((...((((.((....)).)))).(((((.((((........)))).)).))).))))))))))---------------)............................. ( -24.80) >DroSim_CAF1 24183 99 + 1 GUUUGGCCGUUUGUUCGACUUGCAUAAGCCAGUCCGGCGGCAUGGAAAACCCCCCAUUUCAGCCCCA------AA---------------CUCAAACCCAACCCCGUUUUUCAACCCUGC ((((((..(((((...((((.((....)).)))).(((((.((((........)))).)).))).))------))---------------)))))))....................... ( -22.10) >DroEre_CAF1 24040 97 + 1 GUUUGGCCGUUUGUUCGGCUUGCAUAAGCCAGUCCGGCGGCAUG-AAAACCCCCCAUUUCAGCCCCAAACCCA----------------------ACCCAACCCCGUAUUUCAACCCUGC (((((((((..(....(((((....))))).)..))))(((.((-(((........)))))))).)))))...----------------------......................... ( -23.40) >DroYak_CAF1 21582 108 + 1 GUUUG------------GCUUGCAUAAGCCAGUCCGGCGGCAUGGAAAACCCCCCAUUUUAGCCCCAAACCCAAACCCCCCAAACUCUAACCCAAACCCAACCCCGUAUUUCAACCCUGC (((((------------(((.((....)).))...((.(((((((........))))....))))).............))))))................................... ( -17.60) >consensus GUUUGGCCGUUUGUUCGACUUGCAUAAGCCAGUCCGGCGGCAUGGAAAACCCCCCAUUUCAGCCCCAAACCCAAA_______________CUCAAACCCAACCCCGUUUUUCAACCCUGC (((((...........((((.((....)).)))).((.(((.(((((.........))))))))))..........................)))))....................... (-15.60 = -16.08 + 0.48)

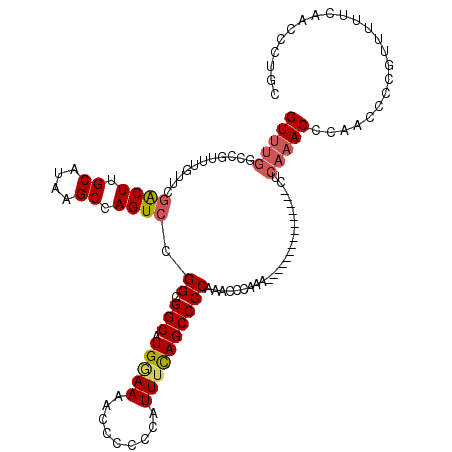
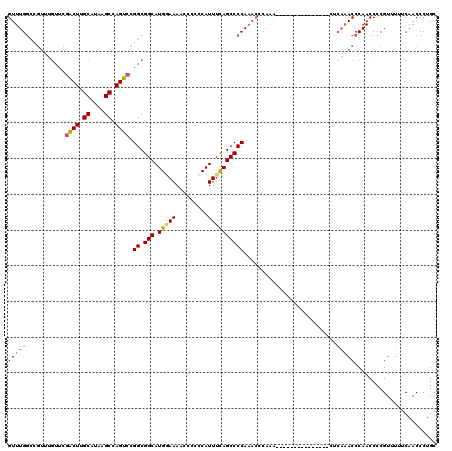
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:12 2006