
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,792,281 – 20,792,429 |
| Length | 148 |
| Max. P | 0.779314 |

| Location | 20,792,281 – 20,792,392 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20792281 111 + 22407834 AUGGAGUUAUCAAAUAAUACAAAAU-UCAAUUGAAGUUUUUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGG-------UCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCU ...(((((..............)))-))......((((((((((......(((((((((((....)))))))))-------))..........((.....)).......)))))))))) ( -23.54) >DroSec_CAF1 15476 111 + 1 AUGGAGUUAUCAAAUAAUGCAAAAU-UCAAUUGAAGUUUUUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGG-------UCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCU ...(((((..((.....))...)))-))......((((((((((......(((((((((((....)))))))))-------))..........((.....)).......)))))))))) ( -23.70) >DroSim_CAF1 19009 111 + 1 AUGGAGUUAUCAAAUAAUACAAAAU-UCAAUUGAAGUUUUUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGG-------UCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCU ...(((((..............)))-))......((((((((((......(((((((((((....)))))))))-------))..........((.....)).......)))))))))) ( -23.54) >DroEre_CAF1 18788 111 + 1 GUGGAGUUAUCAAAUAAUACGAAAUUUCAAUUGAAGUU-UUUACGAGUGCGGCUAGGCAAAUGAUUUCGCUUGG-------UCAUGGAAGAUAUCAAAGUGGAAAAAGAGUGAAAGGCU ....((((.(((........((((((((....))))))-))......(((......)))..)))((((((((..-------((((.((.....))...)))).....)))))))))))) ( -18.60) >DroYak_CAF1 16267 119 + 1 AUGGAGUUAUCAAAUAAUAAAAAAUUUCAAUUGAAGUUUUUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGGUCAUUGGUCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCU .(((((((..............))))))).....((((((((((......(((((((((((....))))))))))).((..((((.((.....))...))))..))...)))))))))) ( -28.54) >consensus AUGGAGUUAUCAAAUAAUACAAAAU_UCAAUUGAAGUUUUUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGG_______UCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCU ..................................((((((((((........(((((((((....))))))))).......((((.((.....))...)))).......)))))))))) (-18.84 = -19.08 + 0.24)



| Location | 20,792,319 – 20,792,429 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20792319 110 + 22407834 UUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGG-------UCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCUUGGAAUGCUCUUUAGUGCUUAGCGCUUUUAAUUGUGU .(((.(((((((((((((((((....)))))))))-------)).............(((((...(((((((..............)))))))..))))).)))))).)))...... ( -31.24) >DroSec_CAF1 15514 110 + 1 UUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGG-------UCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCUUGGAAUGCUCUUUAGCGCUUAGCGCUUUUAAUUGUGU .(((.(((((((((((((((((....)))))))))-------)).............(((((...(((((((..............)))))))..))))).)))))).)))...... ( -33.64) >DroSim_CAF1 19047 110 + 1 UUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGG-------UCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCUUGGAAUGCUCUUUAGCGCUUAGCGCUUUUAAUUGUGU .(((.(((((((((((((((((....)))))))))-------)).............(((((...(((((((..............)))))))..))))).)))))).)))...... ( -33.64) >DroEre_CAF1 18826 103 + 1 UUUACGAGUGCGGCUAGGCAAAUGAUUUCGCUUGG-------UCAUGGAAGAUAUCAAAGUGGAAAAAGAGUGAAAGGCUCGGAAUGCUCUUUA-------CCGCUUUUAAUUGUGU ..(((((....((((((((.((....)).))))))-------))............(((((((..(((((((..............))))))).-------)))))))...))))). ( -25.24) >DroYak_CAF1 16306 109 + 1 UUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGGUCAUUGGUCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCUCG-AAUGCUCUUUA-------GCGCUUUUAAUUGUGU .(((.(((((((((((((((((....))))))))))).....((((.((.....))...))))..(((((((..........-...))))))).-------)))))).)))...... ( -30.62) >consensus UUUACGAGUGCGGCUAGGCAAAUGAUUUUGCUUGG_______UCAUGGAAGAUAUCAAAGUGAAAAAAGAGUGAAAGGCUUGGAAUGCUCUUUAG_GCUUAGCGCUUUUAAUUGUGU .(((.((((((..(((((((((....))))))))).......((((.((.....))...))))..(((((((..............)))))))........)))))).)))...... (-22.48 = -22.72 + 0.24)

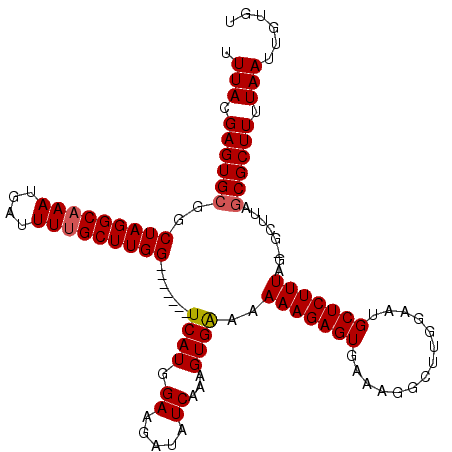
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:03 2006