
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,790,437 – 20,790,536 |
| Length | 99 |
| Max. P | 0.663138 |

| Location | 20,790,437 – 20,790,536 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -24.08 |
| Energy contribution | -26.57 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20790437 99 + 22407834 UCG---------------UCUUGCUGUUCCACCAUCCUCACUGCGUUUGGUAGCCGGAUAGCUCAUAAGUUAAUGGAGAUCUCGUCAUCUCAUCAGUCGCAAACGCAGUGUGAG ...---------------................((..((((((((((.(..((.((.(((((....)))))...(((((......))))).)).))..))))))))))).)). ( -25.60) >DroSec_CAF1 13760 109 + 1 UCGACAGCGGUGGAAUCAUCUUGCUGUUCCACCAUCCUCACUGCGUUUGGUAGCCGGAUAGCUCAUAAGUUAAUGGAGAUCUCGUC-----AUCAGUCGCAAACGCAGUGUGAG ........(((((((.((......))))))))).((..(((((((((((....(((..(((((....))))).))).(((......-----))).....))))))))))).)). ( -35.40) >DroSim_CAF1 16235 109 + 1 UCGACAGUGGUGGAAUCAUCUUGCUGUUCCACCAUCCUCACUGCGUUUGGUAGCCGGAUAGCUCAUAAGUUAAUGGAGAUCUCGUC-----AUCAGUCGCAAACGCAGUGUGAG ......(((((((((.((......))))))))))).(.(((((((((((....(((..(((((....))))).))).(((......-----))).....))))))))))).).. ( -36.70) >DroYak_CAF1 14720 106 + 1 UCGAAA---GUGGAAUCGUCUUGCUGUUCCACCAUCUUCACUGCGUUUGCUACCCGGAUAGCUCAUAACUUAAUGGAGAUCUCGUC-----AUCAGUCGCAAACGCAGUGUGAU ......---(((((((.(.....).))))))).(((..((((((((((((.((...(((..(.(((......)))((....)))..-----))).)).)))))))))))).))) ( -35.00) >consensus UCGACA___GUGGAAUCAUCUUGCUGUUCCACCAUCCUCACUGCGUUUGGUAGCCGGAUAGCUCAUAAGUUAAUGGAGAUCUCGUC_____AUCAGUCGCAAACGCAGUGUGAG ........((((((((.........)))))))).((..(((((((((((....(((..(((((....))))).))).((.((.((......)).)))).))))))))))).)). (-24.08 = -26.57 + 2.50)


| Location | 20,790,437 – 20,790,536 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -21.25 |
| Energy contribution | -23.50 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20790437 99 - 22407834 CUCACACUGCGUUUGCGACUGAUGAGAUGACGAGAUCUCCAUUAACUUAUGAGCUAUCCGGCUACCAAACGCAGUGAGGAUGGUGGAACAGCAAGA---------------CGA ....(((((((((((.(..(((((((((......)))).))))).......((((....)))).))))))))))))....((.((...)).))...---------------... ( -26.90) >DroSec_CAF1 13760 109 - 1 CUCACACUGCGUUUGCGACUGAU-----GACGAGAUCUCCAUUAACUUAUGAGCUAUCCGGCUACCAAACGCAGUGAGGAUGGUGGAACAGCAAGAUGAUUCCACCGCUGUCGA ....(((((((((((.(..((((-----(..(....)..))))).......((((....)))).)))))))))))).((.((((((((((......)).))))))))))..... ( -35.80) >DroSim_CAF1 16235 109 - 1 CUCACACUGCGUUUGCGACUGAU-----GACGAGAUCUCCAUUAACUUAUGAGCUAUCCGGCUACCAAACGCAGUGAGGAUGGUGGAACAGCAAGAUGAUUCCACCACUGUCGA ....(((((((((((.(..((((-----(..(....)..))))).......((((....)))).)))))))))))).((.((((((((((......)).))))))))))..... ( -36.20) >DroYak_CAF1 14720 106 - 1 AUCACACUGCGUUUGCGACUGAU-----GACGAGAUCUCCAUUAAGUUAUGAGCUAUCCGGGUAGCAAACGCAGUGAAGAUGGUGGAACAGCAAGACGAUUCCAC---UUUCGA ....(((((((((((((.((.((-----(((..(((....)))..))))).)))(((....)))))))))))))))..((.(((((((..(.....)..))))))---).)).. ( -37.30) >consensus CUCACACUGCGUUUGCGACUGAU_____GACGAGAUCUCCAUUAACUUAUGAGCUAUCCGGCUACCAAACGCAGUGAGGAUGGUGGAACAGCAAGAUGAUUCCAC___UGUCGA ....(((((((((((........................(((......)))((((....))))..))))))))))).....(((((((...........)))))))........ (-21.25 = -23.50 + 2.25)

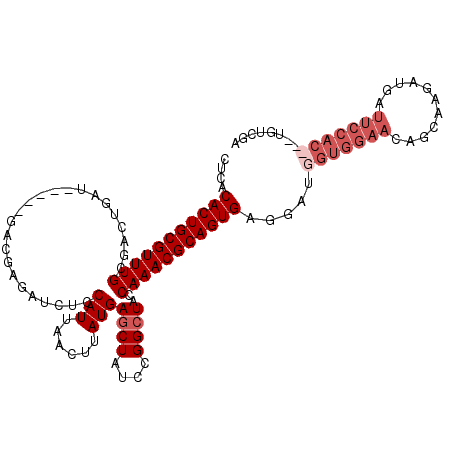
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:01 2006