
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,784,421 – 20,784,604 |
| Length | 183 |
| Max. P | 0.907083 |

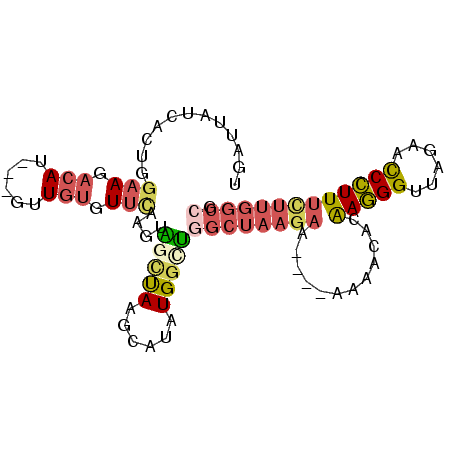
| Location | 20,784,421 – 20,784,513 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 73.93 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -12.90 |
| Energy contribution | -14.02 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20784421 92 - 22407834 UGAUUAUCACUGGAAGACAU---GUUCUGUUCAAGUAGCUAAGCAUAUGGCUGGCUAAGAAA--AAAAACACAAGGGUUAUAACCCUUUUUUGGCCG ........(((((((((...---..))).))).)))(((((......)))))(((((((((.--.........(((((....)))))))))))))). ( -20.90) >DroSec_CAF1 7608 94 - 1 UGACUAUCACUGGAAGACAU---GUUGUCUUCAAGUAGCUAAGCAUAUGGCUGGCUAAGAAACAAAAAACACGAGGGUUAGAACCCUUUCUUGGCCG ........(((.((((((..---...)))))).)))(((((......)))))((((((((............((((((....)))))))))))))). ( -31.50) >DroSim_CAF1 7856 94 - 1 UGACUAUCACUGGAAGACAU---GUUGUCUUUAAGUAGCUAAGCAUAUGGCUGGCUAAGAAACAAAAAACACAAGGGUUAGAACCCUUUCUUGGCCG ........(((.((((((..---...)))))).)))(((((......)))))(((((((((............(((((....)))))))))))))). ( -29.40) >DroEre_CAF1 7767 87 - 1 UGAUUAUCACUGGAAGACAU---GUUGUGUUCCAGAGCUUAAGCAUAUGGCUGGCUAAGAA-------ACACAAGGGUUAGACCCCUUUUUUGGCCG .........((((((.((..---...)).))))))((((.........))))(((((((((-------.....((((......))))))))))))). ( -28.30) >DroYak_CAF1 8153 86 - 1 UGAUUAUUAGAAGAAGACAU---GUUGUGUUCAAGAGGCUAGGC-UAUGGUUGGCUAAGAA-------AUACAAGGGUUAGACUCCUUUCUUGGCGG ............(((.((..---...)).)))....(((((...-..))))).((((((((-------.....((((......)))))))))))).. ( -17.40) >DroAna_CAF1 7737 75 - 1 UGAUUAUAACUCGAAGCCAAUUUUUUGUGU------GCACAAACAUAUGGCC-------------AAAACAUAAAGGUUU---CCUUUUUUUUGCCU ............((((((.....(((((..------..)))))..((((...-------------....))))..)))))---)............. ( -11.60) >consensus UGAUUAUCACUGGAAGACAU___GUUGUGUUCAAGUAGCUAAGCAUAUGGCUGGCUAAGAA____AAAACACAAGGGUUAGAACCCUUUCUUGGCCG ............(((.(((......))).)))....(((((......)))))((((((((............(((((......))))))))))))). (-12.90 = -14.02 + 1.12)


| Location | 20,784,513 – 20,784,604 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -13.76 |
| Consensus MFE | -11.37 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20784513 91 + 22407834 CUUGUUGUUAUUGUUAUAAACACAACGCCACGCAAACAACAAAAACAAUAGCGCGGCUAAGAAUUUCACUCCUCCAACAGCGCAAAAACAA ...((((((.((((.......)))).(((.(((.................))).)))..................)))))).......... ( -14.43) >DroSec_CAF1 7702 91 + 1 CUUGUUGUUAUUGUUAUAAACACAACGCCACGCAAACAACAAAAUCAAAAGCGCGGCCAAGAAUUACACUCCUCCACCAGCGAACAGACAA .((((((...((((.......)))).(((.(((.................))).)))....................))))))........ ( -12.73) >DroSim_CAF1 7950 91 + 1 CUUGUUGUUAUUGUUAUAAACACAACGCCACGCAAACAACAAAAACAAAAGCGCGGCCAAGAAUUACACUCCUCCACCAGCGAACAGACAA .((((((...((((.......)))).(((.(((.................))).)))....................))))))........ ( -12.73) >DroEre_CAF1 7854 91 + 1 CUUGUUGUUAUUGUUAUAAACACAACGCCACGCAAACAACAACAACAAAGACGCGGCUAAAAAUCACACUCCACCAGCAGCGAAAAAACAA .((((((((.(((((...................))))).))))))))...(((.(((.................))).)))......... ( -15.14) >consensus CUUGUUGUUAUUGUUAUAAACACAACGCCACGCAAACAACAAAAACAAAAGCGCGGCCAAGAAUUACACUCCUCCACCAGCGAAAAAACAA .((((((((.((((.......)))).(((.(((.................))).)))..................))))))))........ (-11.37 = -12.30 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:53 2006