
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,761,869 – 20,761,990 |
| Length | 121 |
| Max. P | 0.992291 |

| Location | 20,761,869 – 20,761,962 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -15.35 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20761869 93 + 22407834 -UGUCUGUAGAUUU--UCAGCAUUGCCCGUGU-----UGGGACAUAAAAA-----CUAAUGCCACCUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAU -..(((((....(.--.((((((.....))))-----))..)........-----.....((....(((((((((((...)))))))))))....))))))).... ( -25.40) >DroVir_CAF1 18715 83 + 1 AUGUCUUUAUGUU------CCA----------UUGGGCUGGGCAUAAAAC-----UUAAUGCCACAUGACACAUUGAGAUUUAACGUGUCAAUAUGCACA--AAAU .............------...----------(((.(((((((((.....-----...))))).))((((((.((((...)))).))))))....)).))--)... ( -17.30) >DroPse_CAF1 17790 101 + 1 -UGUCUGUUCAUUUAAUCGUCAUUGUCCAUGU----UUGGGGCAUAUAAAACAGCCUAAUGCCACAUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAU -..(((((.(((...........((((((...----.)))(((((.............))))))))(((((((((((...)))))))))))..))).))))).... ( -25.92) >DroEre_CAF1 13301 78 + 1 -UGUCUGUAU-----------------GGUGC-----UGGGACAUAAAAC-----CUAAUGCCACCUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGGAAAU -..(((((.(-----------------(((..-----((((........)-----)))..))))..(((((((((((...)))))))))))......))))).... ( -24.90) >DroAna_CAF1 13978 94 + 1 -UGUCUGUAGAUUU--UCAGCAUUGGCUGUGU-----UGGGACAUAAAAA----CCUAAUGCCACCUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAU -..(((((.(....--.).((((.((..((((-----((((.........----))))))))..))(((((((((((...)))))))))))..))))))))).... ( -27.80) >DroPer_CAF1 18047 101 + 1 -UGUCUGUUACUUUAAUCGCCAUUGUCCAUGU----UUGGGGCAUAUAAAACAGCCUAAUGCCACAUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAU -..(((((.................((((...----.))))((((((......((.....))....(((((((((((...)))))))))))))))))))))).... ( -25.50) >consensus _UGUCUGUAGAUUU__UC_CCAUUG_CCGUGU_____UGGGACAUAAAAA_____CUAAUGCCACAUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAU ...(((((...............................((.(((.............)))))...(((((((((((...)))))))))))......))))).... (-15.35 = -15.90 + 0.56)

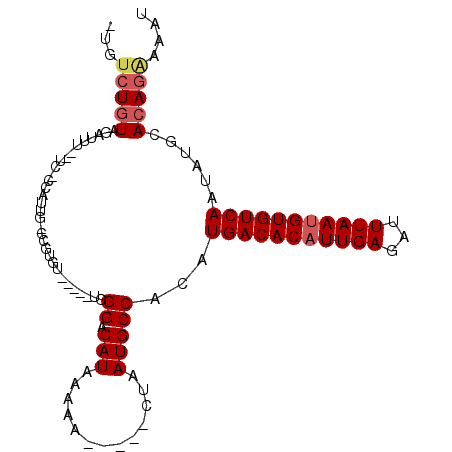

| Location | 20,761,869 – 20,761,962 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20761869 93 - 22407834 AUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAGGUGGCAUUAG-----UUUUUAUGUCCCA-----ACACGGGCAAUGCUGA--AAAUCUACAGACA- ....(((((((...((((((((((.......))))))))))...)).((((-----(.....(((((..-----....)))))..)))))--......)))))..- ( -27.50) >DroVir_CAF1 18715 83 - 1 AUUU--UGUGCAUAUUGACACGUUAAAUCUCAAUGUGUCAUGUGGCAUUAA-----GUUUUAUGCCCAGCCCAA----------UGG------AACAUAAAGACAU ((((--.((((....(((((((((.......)))))))))....)))).))-----))((((((.(((......----------)))------..))))))..... ( -21.20) >DroPse_CAF1 17790 101 - 1 AUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAUGUGGCAUUAGGCUGUUUUAUAUGCCCCAA----ACAUGGACAAUGACGAUUAAAUGAACAGACA- ....(((((.(((..(((((((((.......)))))))))((((((((((((....)))).)))))(((.----...))))))...........))).)))))..- ( -24.00) >DroEre_CAF1 13301 78 - 1 AUUUCCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAGGUGGCAUUAG-----GUUUUAUGUCCCA-----GCACC-----------------AUACAGACA- .......((((....(((((((((.......)))))))))((.(((((...-----.....))))))).-----)))).-----------------.........- ( -22.90) >DroAna_CAF1 13978 94 - 1 AUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAGGUGGCAUUAGG----UUUUUAUGUCCCA-----ACACAGCCAAUGCUGA--AAAUCUACAGACA- ....(((((((...((((((((((.......))))))))))...))).((((----((((..(((....-----)))((((....)))))--)))))))))))..- ( -26.90) >DroPer_CAF1 18047 101 - 1 AUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAUGUGGCAUUAGGCUGUUUUAUAUGCCCCAA----ACAUGGACAAUGGCGAUUAAAGUAACAGACA- ....(((((.(((((.((((((((.......)))))))))))))((....(((..........)))(((.----...)))......))..........)))))..- ( -24.60) >consensus AUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAGGUGGCAUUAG_____UUUUUAUGCCCCA_____ACACGG_CAAUGC_GA__AAAUAUACAGACA_ ....(((((......(((((((((.......)))))))))((.(((((.............)))))))..............................)))))... (-17.08 = -17.52 + 0.44)

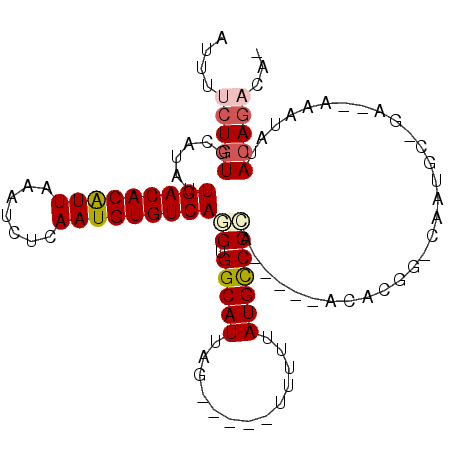
| Location | 20,761,898 – 20,761,990 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.28 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20761898 92 + 22407834 --------UGGGACAUAAAAA-----CUAAUGCCACCUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAUUUAUAUCUAGCC-ACAUAAAUAAUCGAAA- --------..(((.(((((..-----....(((....(((((((((((...)))))))))))....))).......))))).)))....-................- ( -16.62) >DroPse_CAF1 17821 98 + 1 -------UUGGGGCAUAUAAAACAGCCUAAUGCCACAUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAUUUAUAUCGCGCC-ACAUAAAUAAUCGAAA- -------.(((.(((((((((.....((..(((....(((((((((((...)))))))))))....))).))....))))))).)).))-)...............- ( -24.00) >DroEre_CAF1 13315 92 + 1 --------UGGGACAUAAAAC-----CUAAUGCCACCUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGGAAAUUUAUAUCUAGCC-ACAUAAAUAAUCGAAG- --------..(((.(((((.(-----((..(((....(((((((((((...)))))))))))....))).)))...))))).)))....-................- ( -19.50) >DroMoj_CAF1 15711 97 + 1 GGAUGGGGCUGGGCCUAAAAC-----UUAAUGCCACAUGACACAUUGAGAUUUAAUAUGUCAAUAGGCACA--AAAUUUAUAUACGUCCAGCAGA--GAAAGGAAA- (((((((((...)))).....-----....((((...(((((.(((((...))))).)))))...))))..--...........)))))......--.........- ( -22.70) >DroAna_CAF1 14007 94 + 1 --------UGGGACAUAAAAA----CCUAAUGCCACCUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAUUUAUAUCUAGUC-ACAUAAAUAAUAGAAUA --------..(((.(((((..----.....(((....(((((((((((...)))))))))))....))).......))))).)))....-................. ( -17.04) >DroPer_CAF1 18078 98 + 1 -------UUGGGGCAUAUAAAACAGCCUAAUGCCACAUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAUUUAUAUCGCGCC-ACAUAAAUAAUCGAAA- -------.(((.(((((((((.....((..(((....(((((((((((...)))))))))))....))).))....))))))).)).))-)...............- ( -24.00) >consensus ________UGGGACAUAAAAA_____CUAAUGCCACAUGACACAUUGAGAUUUAAUGUGUCAAUAUGCACAGAAAAUUUAUAUCUAGCC_ACAUAAAUAAUCGAAA_ ..............................(((....(((((((((((...)))))))))))....)))...................................... (-13.12 = -13.28 + 0.17)


| Location | 20,761,898 – 20,761,990 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -14.83 |
| Energy contribution | -14.47 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20761898 92 - 22407834 -UUUCGAUUAUUUAUGU-GGCUAGAUAUAAAUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAGGUGGCAUUAG-----UUUUUAUGUCCCA-------- -...............(-((...((((((((..((..((((...((((((((((.......))))))))))...)))).))-----..)))))))))))-------- ( -24.60) >DroPse_CAF1 17821 98 - 1 -UUUCGAUUAUUUAUGU-GGCGCGAUAUAAAUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAUGUGGCAUUAGGCUGUUUUAUAUGCCCCAA------- -...............(-((.((.(((((((...(((((((....(((((((((.......)))))))))....))).))))....))))))))).))).------- ( -26.30) >DroEre_CAF1 13315 92 - 1 -CUUCGAUUAUUUAUGU-GGCUAGAUAUAAAUUUCCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAGGUGGCAUUAG-----GUUUUAUGUCCCA-------- -...............(-((...((((((((...(((((((...((((((((((.......))))))))))...))).)))-----).)))))))))))-------- ( -28.00) >DroMoj_CAF1 15711 97 - 1 -UUUCCUUUC--UCUGCUGGACGUAUAUAAAUUU--UGUGCCUAUUGACAUAUUAAAUCUCAAUGUGUCAUGUGGCAUUAA-----GUUUUAGGCCCAGCCCCAUCC -.........--...(((((.(.((...((((((--.(((((...(((((((((.......)))))))))...))))).))-----)))))).).)))))....... ( -28.90) >DroAna_CAF1 14007 94 - 1 UAUUCUAUUAUUUAUGU-GACUAGAUAUAAAUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAGGUGGCAUUAGG----UUUUUAUGUCCCA-------- .................-.....((((((((...(((((((...((((((((((.......))))))))))...))).))))----..))))))))...-------- ( -25.00) >DroPer_CAF1 18078 98 - 1 -UUUCGAUUAUUUAUGU-GGCGCGAUAUAAAUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAUGUGGCAUUAGGCUGUUUUAUAUGCCCCAA------- -...............(-((.((.(((((((...(((((((....(((((((((.......)))))))))....))).))))....))))))))).))).------- ( -26.30) >consensus _UUUCGAUUAUUUAUGU_GGCUAGAUAUAAAUUUUCUGUGCAUAUUGACACAUUAAAUCUCAAUGUGUCAGGUGGCAUUAG_____UUUUUAUGCCCCA________ ..................(((...(............((((....(((((((((.......)))))))))....))))............)..)))........... (-14.83 = -14.47 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:42 2006