
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,703,882 – 20,703,979 |
| Length | 97 |
| Max. P | 0.697246 |

| Location | 20,703,882 – 20,703,979 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.02 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
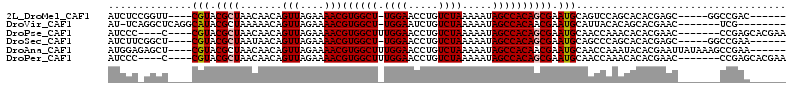
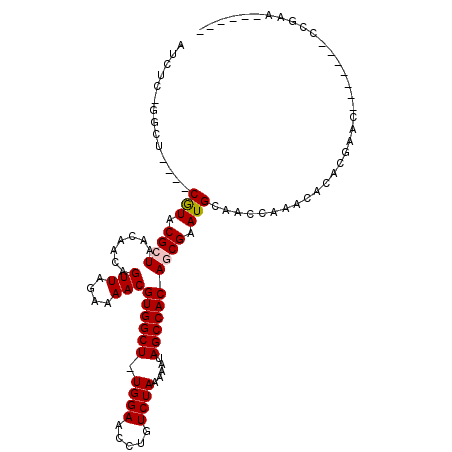
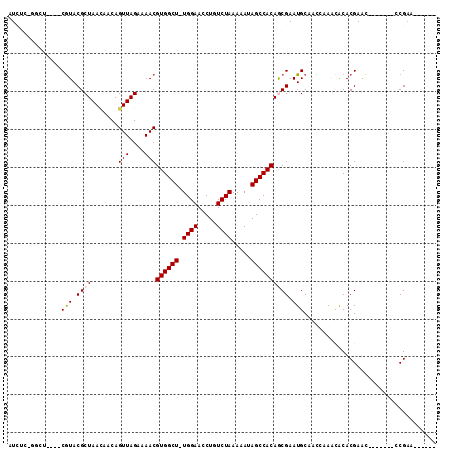
>2L_DroMel_CAF1 20703882 97 - 22407834 AUCUCCGGUU----CGUACGCUAACAACAGUUAGAAAACGUGGCU-UGGAACCUGUCUAAAAAUAGCCACAGCGAAUGCAGUCCAGCACACGAGC-----GGCCGAC------ .....((((.----(((...(((((....)))))....(((((((-.(((..((((.........((....))....)))))))))).)))).))-----)))))..------ ( -26.42) >DroVir_CAF1 3012 96 - 1 AU-UCAGGCUCAGGCAUACGCUAAAAACAGUUAGAAAACGUGGCU-UGGAAUCUGUCUAAAAAUAGCCACAACGAAUGCAUUACACAGCACGAAC-------UCG-------- .(-((..(((...((((.((((((......)))).....((((((-((((.....)))).....))))))..)).)))).......)))..))).-------...-------- ( -19.40) >DroPse_CAF1 33034 98 - 1 AUCCC----C----CGUACGCUAACAACAGUUAGAAAACGUGGCUUUGGAACCUGUCUAAAAAUAGCCACAGCGAAUGCAACCAAACACACGAAC-------CCGAGCACGAA .....----.----(((.((((.......(((....)))(((((((((((.....)))))....))))))))))................((...-------.))...))).. ( -18.70) >DroSec_CAF1 26598 97 - 1 AUCUUCGGCU----CGUACGCUAAUAACAGUUAGAAAACGUGGCU-UGGAACCUGUCUAAAAAUAGCCACAGCGAAUGCAGCCCAGCACACGAGC-----GGCCGAA------ ....(((((.----(((.((((.......(((....)))((((((-((((.....)))).....))))))))))..(((......))).....))-----)))))).------ ( -29.80) >DroAna_CAF1 29043 103 - 1 AUGGAGAGCU----CGUACGCUAACAACAGUUAGAAAACGUGGCUUUGGAACCUGUCUAAAAAUAGCCACAACGAAUGCAACCAAAUACACGAAUUAUAAAGCCGAA------ .(((...(((----(((...(((((....))))).....(((((((((((.....)))))....)))))).))))..))..))).......................------ ( -22.00) >DroPer_CAF1 33000 98 - 1 AUCCC----C----CGUACGCUAACAACAGUUAGAAAACGUGGCUUUGGAACCUGUCUAAAAAUAGCCACAGCGAAUGCAACCAAACACACGAAC-------CCGAGCACGAA .....----.----(((.((((.......(((....)))(((((((((((.....)))))....))))))))))................((...-------.))...))).. ( -18.70) >consensus AUCUC_GGCU____CGUACGCUAACAACAGUUAGAAAACGUGGCU_UGGAACCUGUCUAAAAAUAGCCACAGCGAAUGCAACCAAACACACGAAC_______CCGAA______ ..............(((.((((.......(((....)))((((((.((((.....)))).....)))))))))).)))................................... (-14.82 = -15.02 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:30 2006