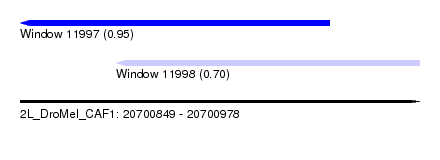
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,700,849 – 20,700,978 |
| Length | 129 |
| Max. P | 0.952939 |

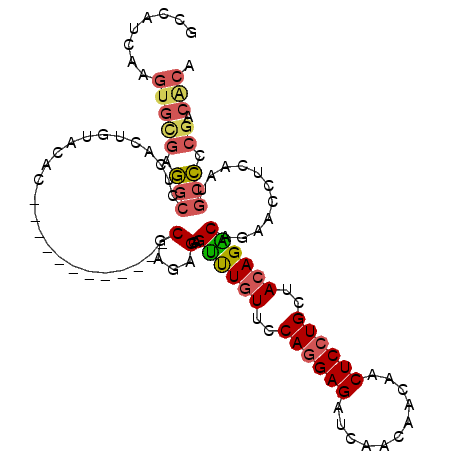
| Location | 20,700,849 – 20,700,949 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -18.06 |
| Energy contribution | -17.42 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
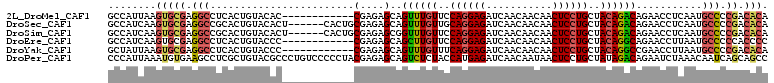
>2L_DroMel_CAF1 20700849 100 - 22407834 -CGAGAGCAGUUUGUUCCAGGAGAUCAACAACAACUCCUGCUACAGACAGAACCUCAAUGCCCCGACACACCAGC---AGUCACACACAUCACACCUGCAACAC -((.(.(((((((((..((((((...........))))))..)))))).(.....)..))).)))........((---((...............))))..... ( -21.96) >DroSec_CAF1 23710 104 - 1 GCGAGAGCAGUUUGUUGCAGGAGAUCAACAACAACUCCUGCUACAGACAGAACCUCAAUGCCCCGACACACCAGCAGCAGGCACACACAUCACACCUGCAACAC ((....)).((((((.(((((((...........))))))).))))))..........(((............)))(((((.............)))))..... ( -30.32) >DroSim_CAF1 15136 104 - 1 GCGAGAGCGGUUUGUUCCAGGAGAUCAACAACAACUCCUGCUACAGACAGAACCUCAAUGCCCCGACACACCAGCAGCAGGCACACACAUCACACCUGCAACAC ((....)).((((((..((((((...........))))))..))))))..........(((............)))(((((.............)))))..... ( -26.82) >DroEre_CAF1 20658 100 - 1 -CGAGAGCAGCUUGUUCCAGGAGAUCAACAACAACUCCUGCUACAGGCAGAACCUUAAUGCCCCCACCCCACAGC---AGCCACACACUUCACACCUGCCACAC -...(.(((((((((..((((((...........))))))..))))))..........(((............))---).................)))).... ( -20.90) >DroYak_CAF1 23961 100 - 1 -CGAGAGCAGUUUGUUUCAGGAGAUCAACAACAACUCCUGCUACAGGCCGAACCUUAAUGCCCCGACACAACAGC---AGCCACACACAUCACACCUGCAACAC -((.(.(((((((((..((((((...........))))))..))))))..........))).)))........((---((...............))))..... ( -19.36) >consensus _CGAGAGCAGUUUGUUCCAGGAGAUCAACAACAACUCCUGCUACAGACAGAACCUCAAUGCCCCGACACACCAGC___AGCCACACACAUCACACCUGCAACAC ..(((..(.((((((..((((((...........))))))..)))))).)...)))................................................ (-18.06 = -17.42 + -0.64)



| Location | 20,700,880 – 20,700,978 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20700880 98 - 22407834 GCCAUUAAGUGCGAGGCCUCACUGUACAC------------CGAGAGCAGUUUGUUCCAGGAGAUCAACAACAACUCCUGCUACAGACAGAACCUCAAUGCCCCGACACA ........(((((.((((((.........------------.)))....((((((..((((((...........))))))..))))))...........))).)).))). ( -27.50) >DroSec_CAF1 23744 104 - 1 GCCAUCAAGUGCGAGGCCGCACUGUACACU------CACUGCGAGAGCAGUUUGUUGCAGGAGAUCAACAACAACUCCUGCUACAGACAGAACCUCAAUGCCCCGACACA ........(((((.(((((((.((......------)).)))).(((..((((((.(((((((...........))))))).)))))).....)))...))).)).))). ( -36.50) >DroSim_CAF1 15170 104 - 1 GCCAUCAAGUGCGAGGCCGCACUGUACACU------CACUGCGAGAGCGGUUUGUUCCAGGAGAUCAACAACAACUCCUGCUACAGACAGAACCUCAAUGCCCCGACACA ........(((((.(((((((.((......------)).)))).(((..((((((..((((((...........))))))..)))))).....)))...))).)).))). ( -34.30) >DroEre_CAF1 20689 98 - 1 GCCAUCAAGUGCGAGGCCUCACUGUACCC------------CGAGAGCAGCUUGUUCCAGGAGAUCAACAACAACUCCUGCUACAGGCAGAACCUUAAUGCCCCCACCCC ........(((.(.((((((.........------------.)))....((((((..((((((...........))))))..))))))...........))).))))... ( -26.10) >DroYak_CAF1 23992 98 - 1 GCUAUUAAGUGCGAGGCCUCACUGUACCC------------CGAGAGCAGUUUGUUUCAGGAGAUCAACAACAACUCCUGCUACAGGCCGAACCUUAAUGCCCCGACACA ((.((((((.(...((((..(((((.(..------------...).))))).(((..((((((...........))))))..)))))))...)))))))))......... ( -27.50) >DroPer_CAF1 29357 110 - 1 CCCAUUAAAUGUGAAGCCUCGCUGUACGCCCUGUCCCCCUACGAGAGCAGUCUCUACCAUGAGAUCAACAAUAACUCCUGCUAUAGACAGAAUCUAAACAAUCAGCAGCC ..(((.....))).......(((((..((.((((......)).)).)).((((.((.((.(((...........))).)).)).))))................))))). ( -16.60) >consensus GCCAUCAAGUGCGAGGCCUCACUGUACAC____________CGAGAGCAGUUUGUUCCAGGAGAUCAACAACAACUCCUGCUACAGACAGAACCUCAAUGCCCCGACACA ........(((((.(((........................(....)..((((((..((((((...........))))))..))))))...........))).)).))). (-18.30 = -18.80 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:28 2006