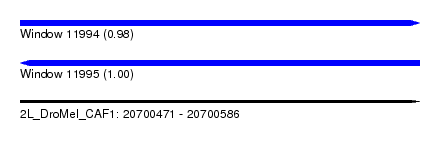
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,700,471 – 20,700,586 |
| Length | 115 |
| Max. P | 0.999917 |

| Location | 20,700,471 – 20,700,586 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.11 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -18.87 |
| Energy contribution | -20.32 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
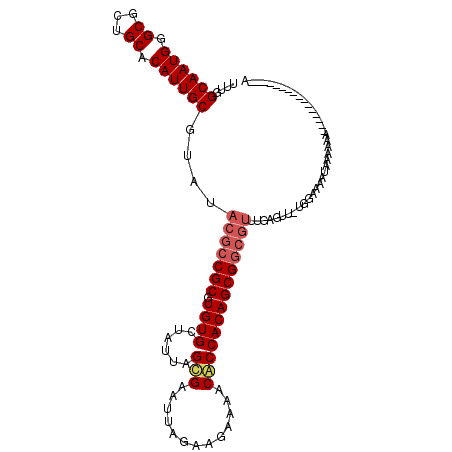
>2L_DroMel_CAF1 20700471 115 + 22407834 UUUGGCAAUGCGCGCUGCACAUUGCGUAUACGCCGCGUGUGCUAUUAGCGAAUUAGAAGAAAACACCACAGCGGCGUUUGAGUU-UGGAAAAUACAAAAAAACAAAAAACGAAAAA ((((((((((.((...)).))))))......(((((((((.((.((((....)))).))...))))....)))))((((...((-((.......)))).))))......))))... ( -28.20) >DroEre_CAF1 20293 87 + 1 UUUGGCAAUGGGCGCUGCACAUUGCGUAUACGACGCGUGUGCGAUUAGCGGAUCAGAAGAAGACGCCACAGCGG-------------AAAAAAGAAAAC----------------A (((.((....((((((((((((.((((.....))))))))))((((....))))......)).))))...)).)-------------))..........----------------. ( -27.30) >DroYak_CAF1 23596 100 + 1 UUUGGCAAUGGGCGCUGCACAUUGCGUAUACGCCGCGUGUGCUCUUAGUGAAUUAGAAGAAAACACCACAGCGGCGUUUGAGUUUUGGGAAAUAUAAAA----------------A ....((((((.((...)).)))))).((.(((((((((((..((((..........))))..))))....))))))).))...................----------------. ( -26.50) >consensus UUUGGCAAUGGGCGCUGCACAUUGCGUAUACGCCGCGUGUGCUAUUAGCGAAUUAGAAGAAAACACCACAGCGGCGUUUGAGUU_UGGAAAAUAAAAAA________________A ....((((((.((...)).))))))....(((((((.((((......(((.............))))))))))))))....................................... (-18.87 = -20.32 + 1.45)

| Location | 20,700,471 – 20,700,586 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.11 |
| Mean single sequence MFE | -27.21 |
| Consensus MFE | -20.96 |
| Energy contribution | -21.63 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.77 |
| SVM decision value | 4.54 |
| SVM RNA-class probability | 0.999917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
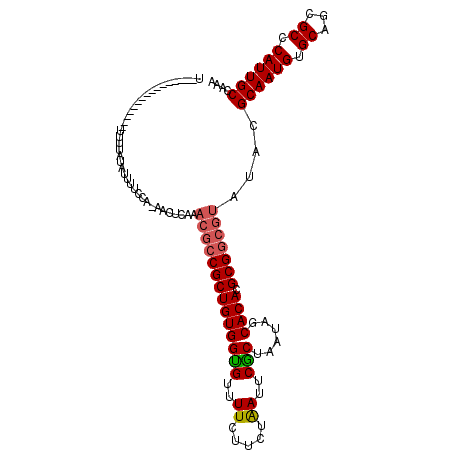
>2L_DroMel_CAF1 20700471 115 - 22407834 UUUUUCGUUUUUUGUUUUUUUGUAUUUUCCA-AACUCAAACGCCGCUGUGGUGUUUUCUUCUAAUUCGCUAAUAGCACACGCGGCGUAUACGCAAUGUGCAGCGCGCAUUGCCAAA ...............................-.......(((((((((((.((((..(.........)..)))).)))).)))))))....(((((((((...))))))))).... ( -32.80) >DroEre_CAF1 20293 87 - 1 U----------------GUUUUCUUUUUU-------------CCGCUGUGGCGUCUUCUUCUGAUCCGCUAAUCGCACACGCGUCGUAUACGCAAUGUGCAGCGCCCAUUGCCAAA .----------------............-------------.((((((((.(((.......))))))).....(((((.((((.....))))..)))))))))............ ( -21.60) >DroYak_CAF1 23596 100 - 1 U----------------UUUUAUAUUUCCCAAAACUCAAACGCCGCUGUGGUGUUUUCUUCUAAUUCACUAAGAGCACACGCGGCGUAUACGCAAUGUGCAGCGCCCAUUGCCAAA .----------------......................(((((((.(((.((((((..............))))))))))))))))....((((((.((...)).)))))).... ( -27.24) >consensus U________________UUUUAUAUUUUCCA_AACUCAAACGCCGCUGUGGUGUUUUCUUCUAAUUCGCUAAUAGCACACGCGGCGUAUACGCAAUGUGCAGCGCCCAUUGCCAAA .......................................((((((((((((((..((.....))..)))......)))).)))))))....((((((.((...)).)))))).... (-20.96 = -21.63 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:26 2006