
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,699,358 – 20,699,523 |
| Length | 165 |
| Max. P | 0.973998 |

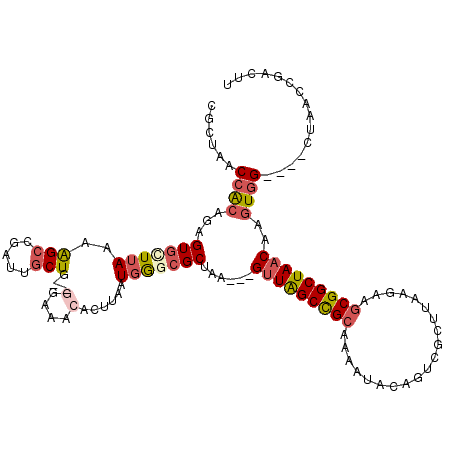
| Location | 20,699,358 – 20,699,469 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20699358 111 + 22407834 CGCUAACCGCAGAGUGCUUAAAAGCUGAUUGCUG--GGAAACACUUAAUGGGCGCUAA---GUUAGCCGCAAAAUACAGUCGCUUAAGAAGCGGCUAACAAGUGG----CUAACCGACUU .((.....))..((((((((..(((.....))).--(....)......))))))))..---(((((((((.......(((((((.....))))))).....))))----)))))...... ( -37.10) >DroSec_CAF1 22311 111 + 1 CACUAACCGCAGAGUGCUUAAAAGCUGAUUGCUG--CGAAACACUUAAUGGGCGCUAA---GUUAGCCGCAAAAUACAGUCGCUUAAGAAGCGGCUAACAAGUGG----CUAACCGACUU ......(((..(((((.((...(((.....))).--..)).)))))..))).......---(((((((((.......(((((((.....))))))).....))))----)))))...... ( -34.60) >DroSim_CAF1 13294 111 + 1 CGCUAACCGCAGAGUGCUUAAAAGCUGAUUGCUG--CGAAACACUUAAUGGGCGCUAA---GUUAGCCGCAAAAUACAGUCGCUUAAGAAGCGGCUAACAAGUGG----CUAACCGACUU .((...(((..(((((.((...(((.....))).--..)).)))))..)))..))...---(((((((((.......(((((((.....))))))).....))))----)))))...... ( -36.50) >DroEre_CAF1 19403 113 + 1 UGCUAGCCACAGAGUGUUUA--AGCCAAGUGCUG--GGCAACACUUAAUGGCCGCCAA---GUUAGCCGCAAAGUACGAACGCUUGGGAAGCGGCUAACAAGUGGCUGGCUAACCGACUU .(((((((((.(((((((..--.(((........--))))))))))............---(((((((((.((((......)))).....)))))))))..))))))))).......... ( -45.90) >DroYak_CAF1 22611 111 + 1 UGCUAGCCACAGAGUGUUUAAAAGCCGAGUGCUU--GGAAACAAUUAAUGGGCGCUAA---GUUAGCUGCAAAACACAGACUCUUAAGAAGCGGCUAACAAGUGG----CUAACCAACUU ...(((((((..((((((((.((((.....))))--(....)......))))))))..---(((((((((....................)))))))))..))))----)))........ ( -35.75) >DroAna_CAF1 24513 104 + 1 AACUAACCACACCGUAUUAAAUGGCCAACUACUAACGAAAACACUCAAUAACCGCUAACUAGCUGGCCG------------GCUUAAGAAGCGGCUAACAACUCG----GUAACCGAUUU ...........((((.((((.((((((.........((......)).......(((....)))))))))------------..))))...))))........(((----(...))))... ( -20.40) >consensus CGCUAACCACAGAGUGCUUAAAAGCCGAUUGCUG__GGAAACACUUAAUGGGCGCUAA___GUUAGCCGCAAAAUACAGUCGCUUAAGAAGCGGCUAACAAGUGG____CUAACCGACUU ......((((...(((((((..(((.....)))...(....)......)))))))......(((((((((....................)))))))))..))))............... (-20.32 = -21.27 + 0.95)

| Location | 20,699,358 – 20,699,469 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -35.43 |
| Consensus MFE | -22.22 |
| Energy contribution | -21.92 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20699358 111 - 22407834 AAGUCGGUUAG----CCACUUGUUAGCCGCUUCUUAAGCGACUGUAUUUUGCGGCUAAC---UUAGCGCCCAUUAAGUGUUUCC--CAGCAAUCAGCUUUUAAGCACUCUGCGGUUAGCG ......(((((----((....(((((((((....................)))))))))---.....((......(((((((..--.(((.....)))...)))))))..))))))))). ( -33.65) >DroSec_CAF1 22311 111 - 1 AAGUCGGUUAG----CCACUUGUUAGCCGCUUCUUAAGCGACUGUAUUUUGCGGCUAAC---UUAGCGCCCAUUAAGUGUUUCG--CAGCAAUCAGCUUUUAAGCACUCUGCGGUUAGUG .((((((((((----(.....)))))))(((.....)))))))..........((((((---..(((((.......))))).((--(((......(((....)))...))))))))))). ( -30.60) >DroSim_CAF1 13294 111 - 1 AAGUCGGUUAG----CCACUUGUUAGCCGCUUCUUAAGCGACUGUAUUUUGCGGCUAAC---UUAGCGCCCAUUAAGUGUUUCG--CAGCAAUCAGCUUUUAAGCACUCUGCGGUUAGCG ((((.((....----))))))(((((((((..((((((.(.(((.(((((((((...((---((((......))))))...)))--))).))))))).))))))......))))))))). ( -34.40) >DroEre_CAF1 19403 113 - 1 AAGUCGGUUAGCCAGCCACUUGUUAGCCGCUUCCCAAGCGUUCGUACUUUGCGGCUAAC---UUGGCGGCCAUUAAGUGUUGCC--CAGCACUUGGCU--UAAACACUCUGUGGCUAGCA ......((((((((((((...(((((((((.......((....)).....)))))))))---.))))(((((...((((((...--.)))))))))))--...........)))))))). ( -46.80) >DroYak_CAF1 22611 111 - 1 AAGUUGGUUAG----CCACUUGUUAGCCGCUUCUUAAGAGUCUGUGUUUUGCAGCUAAC---UUAGCGCCCAUUAAUUGUUUCC--AAGCACUCGGCUUUUAAACACUCUGUGGCUAGCA ((((.((....----))))))(((((((((...(((((((((.((((((.((.((((..---.)))))).((.....)).....--))))))..))))))))).......))))))))). ( -32.80) >DroAna_CAF1 24513 104 - 1 AAAUCGGUUAC----CGAGUUGUUAGCCGCUUCUUAAGC------------CGGCCAGCUAGUUAGCGGUUAUUGAGUGUUUUCGUUAGUAGUUGGCCAUUUAAUACGGUGUGGUUAGUU ...((((...)----)))....((((((((.(((((((.------------.(((((((((.(((((((..((.....))..)))))))))))))))).)))))...)).)))))))).. ( -34.30) >consensus AAGUCGGUUAG____CCACUUGUUAGCCGCUUCUUAAGCGACUGUAUUUUGCGGCUAAC___UUAGCGCCCAUUAAGUGUUUCC__CAGCAAUCAGCUUUUAAACACUCUGCGGUUAGCG ((((.((........))))))(((((((((.......................(((((....))))).........((((((.....(((.....)))...))))))...))))))))). (-22.22 = -21.92 + -0.30)
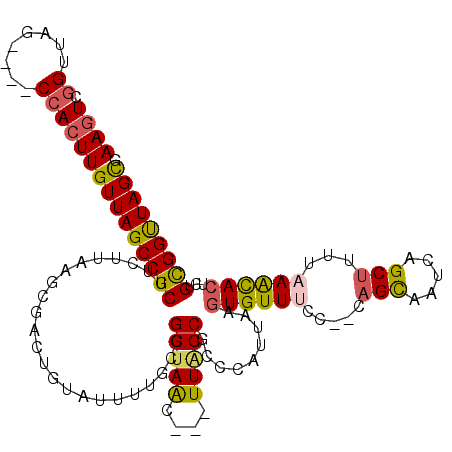
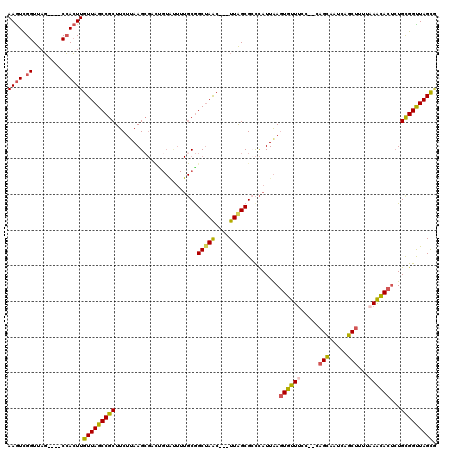
| Location | 20,699,433 – 20,699,523 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.97 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
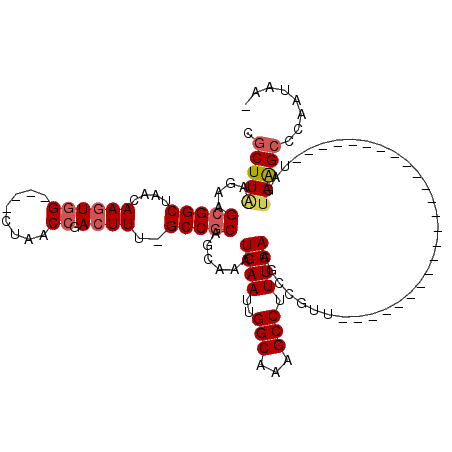
>2L_DroMel_CAF1 20699433 90 + 22407834 CGCUUAAGAAGCGGCUAACAAGUGG----CUAACCGACUUU-GCCGCAGCGAAUGAAUUGGCAAAGCCUUUCAAGCCGUU------------------------UAGUAAGCCCAAUAA- .(((((..((((((((.....((((----(...........-)))))......((((..(((...))).)))))))))))------------------------)..))))).......- ( -32.30) >DroSec_CAF1 22386 90 + 1 CGCUUAAGAAGCGGCUAACAAGUGG----CUAACCGACUUU-GCCGCAGCGAAUGAAUUGGCAAAGCCUUUCAAGCCGUU------------------------UAGUAAGCCCAAUAA- .(((((..((((((((.....((((----(...........-)))))......((((..(((...))).)))))))))))------------------------)..))))).......- ( -32.30) >DroSim_CAF1 13369 90 + 1 CGCUUAAGAAGCGGCUAACAAGUGG----CUAACCGACUUU-GCCGCAGCGAAUGAAUUGGCAAAGCCUUUCAAGCCGUU------------------------UAGUAAGCCCAAUAA- .(((((..((((((((.....((((----(...........-)))))......((((..(((...))).)))))))))))------------------------)..))))).......- ( -32.30) >DroEre_CAF1 19476 94 + 1 CGCUUGGGAAGCGGCUAACAAGUGGCUGGCUAACCGACUUU-GCCGCAGCAAAUGAAUUGGCAAAGCCUUUCAAGCCGUU------------------------UAGUAAGCUCAAUAA- .(((((..((((((((.....((((((((....))).....-)))))......((((..(((...))).)))))))))))------------------------)..))))).......- ( -32.20) >DroYak_CAF1 22686 90 + 1 CUCUUAAGAAGCGGCUAACAAGUGG----CUAACCAACUUU-GCCGCAGCAAAUGAAUUGGCAAAGCCUUUCAAGGCGUU------------------------UAGUAAGCUCAAUAA- ..((((..(((((.((.....((((----(...........-)))))......((((..(((...))).)))))).))))------------------------)..))))........- ( -23.70) >DroAna_CAF1 24582 115 + 1 -GCUUAAGAAGCGGCUAACAACUCG----GUAACCGAUUUUGGCCACAACAAAUGAAUUGGCAAAGCCUUUCAAGCCGUUACCAAUUCCACACCAACGGCGAUUAAGAAGGCUCAAUAAA -(((.....)))((((((....(((----(...))))..))))))...........((((....(((((((...((((((..............))))))......)))))))))))... ( -28.34) >consensus CGCUUAAGAAGCGGCUAACAAGUGG____CUAACCGACUUU_GCCGCAGCAAAUGAAUUGGCAAAGCCUUUCAAGCCGUU________________________UAGUAAGCCCAAUAA_ .(((((....(((((....((((((........)).))))..)))))......((((..(((...))).))))..................................)))))........ (-17.38 = -17.97 + 0.59)

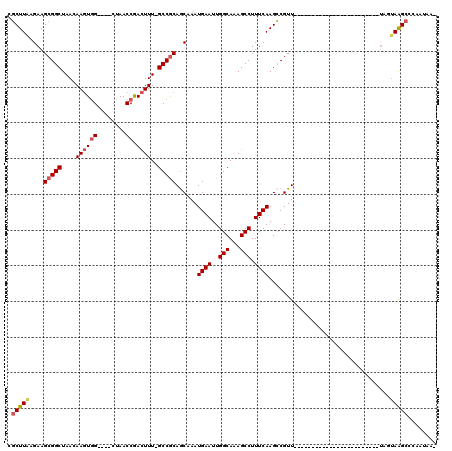
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:24 2006