
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
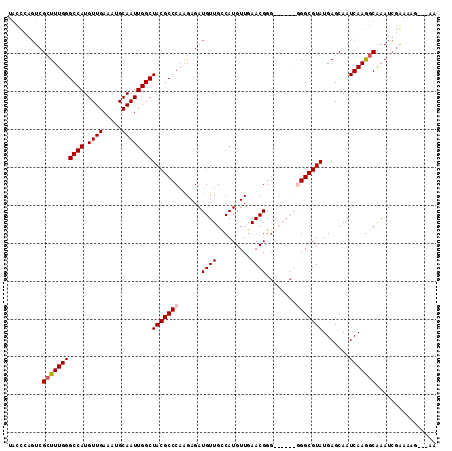
| Location | 20,698,379 – 20,698,481 |
| Length | 102 |
| Max. P | 0.817596 |

| Location | 20,698,379 – 20,698,481 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.94 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
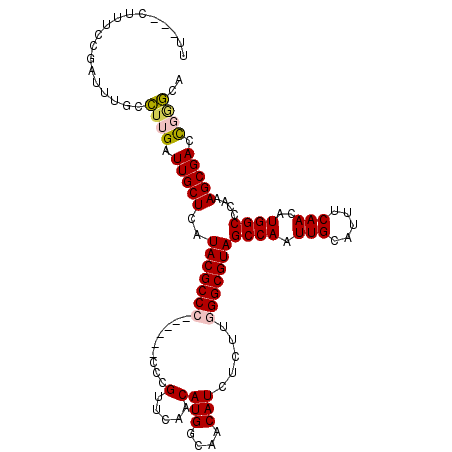
>2L_DroMel_CAF1 20698379 102 + 22407834 UGCCGGGUCGCUUUGGGCCAUGUUGAAAUGCAAUUGGCUACGCCCAAGAGAUGUUGCCAUGUUGAACGG--------GGCGUAUGAGCAAUCAAGGCAAAUCGGAAAG---AA ..((((...(((((((((((.((((.....))))))))(((((((..((.(((....))).)).....)--------)))))).......)))))))...))))....---.. ( -34.60) >DroSec_CAF1 21361 104 + 1 UGCCCGGUCGCUUUGGGCCAUGUUGAAAUGCAAUUGGCUACGCCCAAGAGAUGUUGCCAUGUUGAACGGG------CGGCGUAUGAGCAAUCAAGGCAAAUCGGAAAG---AA ...(((((.(((((((((((.((((.....)))))))))..((.((....((((((((.((.....))))------)))))).)).))...))))))..)))))....---.. ( -34.70) >DroYak_CAF1 21578 110 + 1 UACUCAGUCGCUUUGGGCCAUGUUGAAAUGCAAUUGGCUACGCCCCAGAGAUGUUGUCAUGUUGAACGGGGCAACGGGGCGUAUGAGCAAUCAAGGCGAAUCGAAAAG---AA ...((..(((((((((((((.((((.....))))))))((((((((......((((((.((.....)).)))))))))))))).......)))))))))...))....---.. ( -40.00) >DroAna_CAF1 23676 111 + 1 UAU--AUUCGCUUUGGGCCAUGUUGAAAUGCAAUUGGCUACGCCCAGGAGAUGUUGCCAUGUUCAACGGGGCAACGGGGCGUAUGAGCAAUCAAAUCAAAUGAAAGAGUUAAA ...--.((((.(((((((((.((((.....))))))))(((((((......(((((((.((.....)).))))))))))))))(((....)))..)))))))))......... ( -34.00) >consensus UACCCAGUCGCUUUGGGCCAUGUUGAAAUGCAAUUGGCUACGCCCAAGAGAUGUUGCCAUGUUGAACGGG______GGGCGUAUGAGCAAUCAAGGCAAAUCGAAAAG___AA .........(((((((((((.((((.....))))))))(((((((......((((.........))))........))))))).......)))))))................ (-23.38 = -23.94 + 0.56)


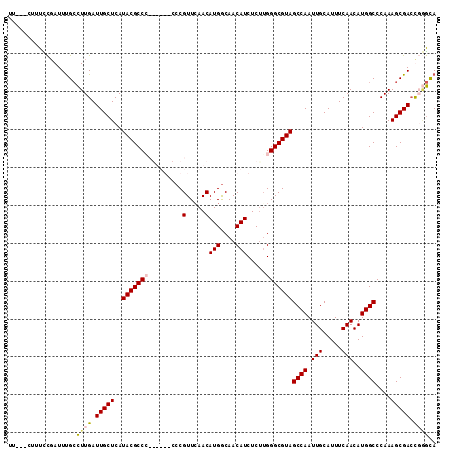
| Location | 20,698,379 – 20,698,481 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -18.83 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20698379 102 - 22407834 UU---CUUUCCGAUUUGCCUUGAUUGCUCAUACGCC--------CCGUUCAACAUGGCAACAUCUCUUGGGCGUAGCCAAUUGCAUUUCAACAUGGCCCAAAGCGACCCGGCA ..---....(((..((((.(((........((((((--------(........(((....))).....)))))))((((.(((.....)))..)))).))).))))..))).. ( -27.62) >DroSec_CAF1 21361 104 - 1 UU---CUUUCCGAUUUGCCUUGAUUGCUCAUACGCCG------CCCGUUCAACAUGGCAACAUCUCUUGGGCGUAGCCAAUUGCAUUUCAACAUGGCCCAAAGCGACCGGGCA ..---..........(((((.(.(((((.....((((------((((......(((....)))....)))))((((....))))..........)))....)))))).))))) ( -28.60) >DroYak_CAF1 21578 110 - 1 UU---CUUUUCGAUUCGCCUUGAUUGCUCAUACGCCCCGUUGCCCCGUUCAACAUGACAACAUCUCUGGGGCGUAGCCAAUUGCAUUUCAACAUGGCCCAAAGCGACUGAGUA ..---...((((..((((.(((........((((((((((((.......))))(((....)))....))))))))((((.(((.....)))..)))).))).)))).)))).. ( -29.30) >DroAna_CAF1 23676 111 - 1 UUUAACUCUUUCAUUUGAUUUGAUUGCUCAUACGCCCCGUUGCCCCGUUGAACAUGGCAACAUCUCCUGGGCGUAGCCAAUUGCAUUUCAACAUGGCCCAAAGCGAAU--AUA ............(((((.((((........(((((((.((((((..(.....)..)))))).......)))))))((((.(((.....)))..)))).)))).)))))--... ( -26.60) >consensus UU___CUUUCCGAUUUGCCUUGAUUGCUCAUACGCCC______CCCGUUCAACAUGGCAACAUCUCUUGGGCGUAGCCAAUUGCAUUUCAACAUGGCCCAAAGCGACCGGGCA ..................((((.(((((..(((((((.........(.....)(((....))).....)))))))((((.(((.....)))..))))....))))).)))).. (-18.83 = -19.32 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:22 2006