
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,692,278 – 20,692,388 |
| Length | 110 |
| Max. P | 0.707153 |

| Location | 20,692,278 – 20,692,388 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.10 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
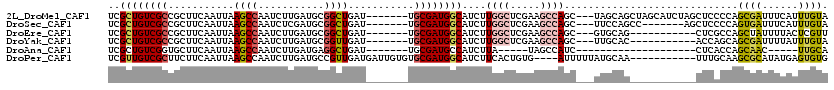
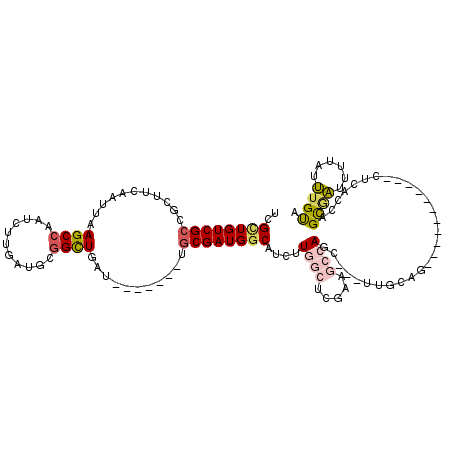
>2L_DroMel_CAF1 20692278 110 + 22407834 UCGCUGUCGCCGCUUCAAUUAAGCCAAUCUUGAUGCGGCUGAU-------UGCGAUGGCAUCUUGGCUCGAAGCCAGC---UAGCAGCUAGCAUCUAGCUCCCCAGCGAUUUCAUUUGUA ((((.((((((((.((((...........)))).))))).)))-------.))))(((.(((.((((.....))))((---(.(.((((((...)))))).)..)))))).)))...... ( -37.30) >DroSec_CAF1 15465 103 + 1 UCGCUGUCGCCGCUUCAAUUAAGCCAAUCUCGAUGCGGCUGAU-------UGCGAUGGCAUCUUGGCUCGAAGCCAGC---UUCCAGCC-------AGCUCCCCAGUGAUUUCAUUUGUA ((((((..((((...((((((.(((.((....))..)))))))-------))...))))...((((((.((((....)---))).))))-------)).....))))))........... ( -33.70) >DroEre_CAF1 13358 99 + 1 UCGCUGUCGCCGCUUCAAUUAAGCCAAUCUUGAUGCGGCUGAU-------UGCGAUGGCAUCUUGGCUCGAAGCCAGC---GUGCAG-----------CUCGCCAGCUAUUUUACUCGUU ..((((..(((((.((((...........)))).)))))....-------.((((..((((.(((((.....))))).---))))..-----------.))))))))............. ( -35.20) >DroYak_CAF1 15571 99 + 1 UCGCUGUCGCCGCUUCAAUUAAGCCAAUCUUGAUGCGGUUGAU-------UGCGAUGGCAUCUUGGCUCGAAGCCAGC---UUGCAC-----------ACCAGCAGCGAUUUUAUUUGUA (((((((.((.(((((.....((((((....(((((.((((..-------..)))).))))))))))).)))))..))---.((...-----------..)))))))))........... ( -32.30) >DroAna_CAF1 18000 83 + 1 UCGCUGUCGGUGCUUCAAUUAAGCCAAUCUUGAUGAGGCUGAU-------UGCGAUGCCAUCUUA-----UAGCCAUC--------------------CUCACCAGCAAC-----UUGCA ((((.((((..((((((.(((((.....)))))))))))))))-------.))))..........-----........--------------------.......((...-----..)). ( -18.70) >DroPer_CAF1 19529 105 + 1 UCGUUGUCGCUUCUUCAAUUAAGCCAAUCUUGAUGCCGUUGAUGAUUGUGUGCGAUGGCAUCUUCACUGUG----AUUUUUAUGCAA-----------UUUGCAAGCGCAUAUGAGUGUG ((((((.((((....................((((((((((((......)).)))))))))).........----.......(((..-----------...))))))))).))))..... ( -24.10) >consensus UCGCUGUCGCCGCUUCAAUUAAGCCAAUCUUGAUGCGGCUGAU_______UGCGAUGGCAUCUUGGCUCGAAGCCAGC___UUGCAG___________CUCACCAGCGAUUUUAUUUGUA ..((((((((...........((((...........))))...........))))))))....((((.....)))).............................((((......)))). (-15.61 = -16.28 + 0.67)
| Location | 20,692,278 – 20,692,388 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.10 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -13.40 |
| Energy contribution | -16.42 |
| Covariance contribution | 3.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
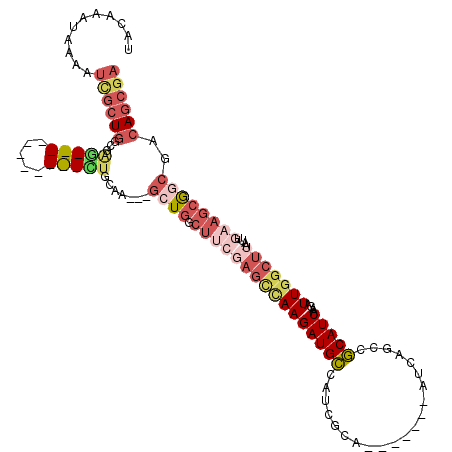
>2L_DroMel_CAF1 20692278 110 - 22407834 UACAAAUGAAAUCGCUGGGGAGCUAGAUGCUAGCUGCUA---GCUGGCUUCGAGCCAAGAUGCCAUCGCA-------AUCAGCCGCAUCAAGAUUGGCUUAAUUGAAGCGGCGACAGCGA ...........((((((((.((((((...)))))).)).---((((.((((((((((((((((....((.-------....)).)))))....)))))))....))))))))..)))))) ( -42.10) >DroSec_CAF1 15465 103 - 1 UACAAAUGAAAUCACUGGGGAGCU-------GGCUGGAA---GCUGGCUUCGAGCCAAGAUGCCAUCGCA-------AUCAGCCGCAUCGAGAUUGGCUUAAUUGAAGCGGCGACAGCGA .................((.(..(-------((((.(((---(....)))).)))))...).)).((((.-------.((.(((((.((((...........)))).)))))))..)))) ( -35.70) >DroEre_CAF1 13358 99 - 1 AACGAGUAAAAUAGCUGGCGAG-----------CUGCAC---GCUGGCUUCGAGCCAAGAUGCCAUCGCA-------AUCAGCCGCAUCAAGAUUGGCUUAAUUGAAGCGGCGACAGCGA .............((((((((.-----------..((((---..((((.....)))).).)))..)))).-------....(((((.((((...........)))).)))))..)))).. ( -35.30) >DroYak_CAF1 15571 99 - 1 UACAAAUAAAAUCGCUGCUGGU-----------GUGCAA---GCUGGCUUCGAGCCAAGAUGCCAUCGCA-------AUCAACCGCAUCAAGAUUGGCUUAAUUGAAGCGGCGACAGCGA ...........(((((((.(((-----------(.(((.---..((((.....))))...)))))))...-------.....((((.((((...........)))).)))).).)))))) ( -30.70) >DroAna_CAF1 18000 83 - 1 UGCAA-----GUUGCUGGUGAG--------------------GAUGGCUA-----UAAGAUGGCAUCGCA-------AUCAGCCUCAUCAAGAUUGGCUUAAUUGAAGCACCGACAGCGA .....-----((((.(((((..--------------------((((.(..-----......).)))).((-------((.((((...........))))..))))...))))).)))).. ( -23.10) >DroPer_CAF1 19529 105 - 1 CACACUCAUAUGCGCUUGCAAA-----------UUGCAUAAAAAU----CACAGUGAAGAUGCCAUCGCACACAAUCAUCAACGGCAUCAAGAUUGGCUUAAUUGAAGAAGCGACAACGA ..........(((((((((...-----------..))........----..((((...((((((...................))))))...))))............))))).)).... ( -18.21) >consensus UACAAAUAAAAUCGCUGGCGAG___________CUGCAA___GCUGGCUUCGAGCCAAGAUGCCAUCGCA_______AUCAGCCGCAUCAAGAUUGGCUUAAUUGAAGCGGCGACAGCGA ...........((((((...(((((.....))))).......((((.((((((((((((((((.....................)))))....)))))))....))))))))..)))))) (-13.40 = -16.42 + 3.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:18 2006