
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,687,163 – 20,687,276 |
| Length | 113 |
| Max. P | 0.959812 |

| Location | 20,687,163 – 20,687,276 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.92 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -13.56 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
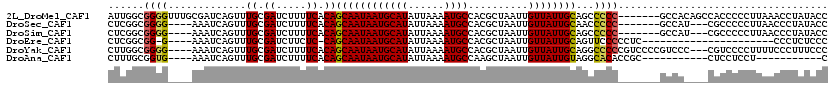
>2L_DroMel_CAF1 20687163 113 - 22407834 AUUGGCGGGGUUUGCGAUCAGUUUGCGAUCUUUUCACAGCAAUAAUGCAUAUUAAAAUGCCACGCUAAUUGUUAUUGCAGCCCCC-------GCCACAGCCACCCCCUUAAACCUAUACC ..((((((((.((((((((((((.((((.....))...........((((......))))...)).)))))..))))))).))))-------))))........................ ( -33.90) >DroSec_CAF1 10565 106 - 1 CUCGGCGGGG----AAAUCAGUUUGCGAUCUUUUCACAGCAAUAAUGCAUAUUAAAAUGCCACGCUAAUUGUUAUUGCAACCCCC-------GCCAU---CGCCCCCUUAACCCUAUACC ...(((((((----.........((.((.....)).))((((((((((((......))))..........))))))))...))))-------)))..---.................... ( -28.00) >DroSim_CAF1 6684 106 - 1 CUCGGCGGGG----AAAUCAGUUUGCGAUCUUUUCACAGCAAUAAUGCAUAUUAAAAUGCCACGCUAAUUGUUAUUGCAGCCCCC-------GCCAU---CGCCCCCUUAACCCUAUACC ...(((((((----.........((.((.....)).))((((((((((((......))))..........))))))))...))))-------)))..---.................... ( -28.00) >DroEre_CAF1 8852 92 - 1 CUCGGCGG-G----AAAUCAGUUUGCGAUCUUCUC-CAGCAAUAAUGCAUAUUAAAAUGCCACGCUAAUUGUUAUUGCAGUUCCCCCUC----------------------CCCUCUCCC ...((.((-(----((.......((.((.....))-))((((((((((((......))))..........))))))))..)))))))..----------------------......... ( -18.60) >DroYak_CAF1 10516 113 - 1 CUUGGCGGGG----AAAUCAGUUUGCGAUCUUUUCACAGCAAUAAUGCAUAUUAAAAUGCCACGCUAAUUGUUAUUGCAGGCCCCCGUCCCCGUCCC---CGUCCCCUUUUCCCUUUCCC ...(((((((----(.....(((((((((........(((......((((......))))...))).......))))))))).....))))))))..---.................... ( -28.76) >DroAna_CAF1 11016 94 - 1 CUUUGCGGUG----AAAUCAGUUUGCGAUCUUUUCACAGCAAUAAUGCAUAUUAAAAUGCCAAGCUAAUUGUUAUUGUAGGCACACCGC-----------CUCCUCCU-----------C ....((((((----.........((.((.....)).))((((((((((((......))))..........)))))))).....))))))-----------........-----------. ( -19.10) >consensus CUCGGCGGGG____AAAUCAGUUUGCGAUCUUUUCACAGCAAUAAUGCAUAUUAAAAUGCCACGCUAAUUGUUAUUGCAGCCCCC_______GCCA____CGCCCCCUUAACCCUAUACC ......((((.............((.((.....)).))((((((((((((......))))..........))))))))...))))................................... (-13.56 = -13.40 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:15 2006