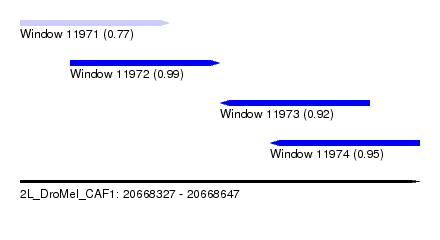
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,668,327 – 20,668,647 |
| Length | 320 |
| Max. P | 0.989825 |

| Location | 20,668,327 – 20,668,447 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -34.30 |
| Energy contribution | -34.38 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20668327 120 + 22407834 GUCUGCAGACCUUGGUUGUCUUGGAUUAUGGUCACCAACUGCUCCUCCAGCUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCACGUUUUAACACCUCCAUUUCGCUGAGCAGCUUUACC (((((.((((.......)))))))))...(((((((....(((.....)))..)))).)))...((((((((((...(((((...((......))....)))))))))))))))...... ( -32.20) >DroSec_CAF1 21175 120 + 1 GUCUGCAGACCUUGGUUGUCUUGGAUUAUGGUCACCAACUGCUCCUCCAGUUCGGUGCACCGCUGCUGUUCGGCCACGAAGUCUCGUUUCAACACCUCCAUUUCGCUGAGCAGCUUUACC (((((.((((.......)))))))))...((((((((((((......))))).)))).)))...((((((((((...(((((...((......))....)))))))))))))))...... ( -35.40) >DroSim_CAF1 21501 120 + 1 GUCUGCAGACCUUGGUUGUCUUGGAUUAUGGUCACCAACUGCUCCUCCAGUUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCUCGUUUCAACACCUCCAUUUCGCUGAGCAGCUUUACC (((((.((((.......)))))))))...((((((((((((......))))).)))).)))...((((((((((...(((((...((......))....)))))))))))))))...... ( -35.40) >DroEre_CAF1 20811 120 + 1 CUCUGCAGACCUUGGUUGUCUUGGAUUAUGGUCACCAGCUGCUCCUCCAGCUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCACGUUUCAACACCUCCAUUUCGCUGAGCAGCUUCACC .((((.((((.......))))))))....((((((((((((......))))).)))).)))(..((((((((((...(((((...((......))....)))))))))))))))..)... ( -37.50) >DroYak_CAF1 20741 120 + 1 CUCUGCAGACCUUGGUUGUCUUGGAUUAUGGUCACCAGCUGCUCCUCCAGCUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCACGUUUCAACACCUCCAUUUCGCUGAUCAGCUUUACC .......(((((((((((...(((....(((((((((((((......))))).)))).))))...)))..))))))))..))).....................(((....)))...... ( -33.40) >consensus GUCUGCAGACCUUGGUUGUCUUGGAUUAUGGUCACCAACUGCUCCUCCAGCUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCACGUUUCAACACCUCCAUUUCGCUGAGCAGCUUUACC .((((.((((.......))))))))....((((((((((((......))))).)))).)))...((((((((((...(((((...((......))....)))))))))))))))...... (-34.30 = -34.38 + 0.08)



| Location | 20,668,367 – 20,668,487 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -31.14 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20668367 120 + 22407834 GCUCCUCCAGCUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCACGUUUUAACACCUCCAUUUCGCUGAGCAGCUUUACCAAUUCCUCGUUGUCUUCACCCUUAACGGCGGUCCUCUCGC ((.((........)).))(((((.((((((((((...(((((...((......))....))))))))))))))).............(((((..........))))))))))........ ( -31.60) >DroSec_CAF1 21215 120 + 1 GCUCCUCCAGUUCGGUGCACCGCUGCUGUUCGGCCACGAAGUCUCGUUUCAACACCUCCAUUUCGCUGAGCAGCUUUACCAAUUCCUCGUUAUCUUCACCCUUAACGGCGGUCCUGUCGC ((.((........)).))(((((.((((((((((...(((((...((......))....))))))))))))))).............(((((..........))))))))))........ ( -31.90) >DroSim_CAF1 21541 120 + 1 GCUCCUCCAGUUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCUCGUUUCAACACCUCCAUUUCGCUGAGCAGCUUUACCAAUUCCUCGUUAUCUUCACCCUUAACGGCGGUCCUGUCGC ((.((........)).))(((((.((((((((((...(((((...((......))....))))))))))))))).............(((((..........))))))))))........ ( -31.90) >DroEre_CAF1 20851 120 + 1 GCUCCUCCAGCUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCACGUUUCAACACCUCCAUUUCGCUGAGCAGCUUCACCAAUUCCUCGUUGUCCUCACCCUUAACGGCGGUCCGCUCAC (((.....)))..((((.(((((.((((((((((...(((((...((......))....))))))))))))))).............(((((..........)))))))))).))))... ( -33.20) >DroYak_CAF1 20781 120 + 1 GCUCCUCCAGCUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCACGUUUCAACACCUCCAUUUCGCUGAUCAGCUUUACCAAUUCCUCGUUGUCCUCACCCUUAACGGCGGUCCUCUCAC ((.((........)).))(((((.((((.(((((...(((((...((......))....)))))))))).)))).............(((((..........))))))))))........ ( -27.50) >consensus GCUCCUCCAGCUCGGUGCACCGCUGCUGUUCGGCCAAGAAGUCACGUUUCAACACCUCCAUUUCGCUGAGCAGCUUUACCAAUUCCUCGUUGUCUUCACCCUUAACGGCGGUCCUCUCGC ((.((........)).))(((((.((((((((((...(((((...((......))....))))))))))))))).............(((((..........))))))))))........ (-31.14 = -31.10 + -0.04)



| Location | 20,668,487 – 20,668,607 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -32.16 |
| Energy contribution | -32.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20668487 120 - 22407834 CCAGUCACAGCAUUCAAUCGCUGUGCAAGGAGCAGAGUGUUGAGUUCUCCAAACUAGAUGUUAUGGCCAUAGCGAACUCGACAGGAUUGGAGGAUAUCUCCUUCAGUAAUGCCACCAUGU ...(.((((((........)))))))..((.(((...((((((((((..(((((.....))).))((....))))))))))))..(((((((((....)))))))))..)))..)).... ( -39.10) >DroSec_CAF1 21335 120 - 1 CCAGUCACAGCAUUCAAUCCCUGUGCAAGGAGCAGAGUGUGGAGUUCUCCAAACUAGAUGUUAUGGCCAUAGCGAACUCGACAGGAUUGGAGGAUAUCUCCUUCAGUAAUGCAACCAUGU .........(((((((((((((.(((.....))).))(((.((((((..(((((.....))).))((....)))))))).))))))))((((((....)))))).).)))))........ ( -35.20) >DroSim_CAF1 21661 120 - 1 CCAGUCACAGCAUUCAAUCCCUGUGCAAGGAGCAGAGUGUGGAGUUCUCCAAACUAGAUGUUAUGGCCAUAGCGAACUCGACAGGAUUGGAGGAUAUCUCCUUCAGUAAUGCAACCAUGU .........(((((((((((((.(((.....))).))(((.((((((..(((((.....))).))((....)))))))).))))))))((((((....)))))).).)))))........ ( -35.20) >DroEre_CAF1 20971 120 - 1 CCAGUCAUAGCAUUCAAUCGCUGUGCAAGGAGCAGAGUGUGGAGUUCUCCAAACUAGAUGUCAUGGCCAUGGCGAACUCGACCGGAUUGGAGGAUCUCUCCUUCAGUAAUGCCACCAUGU ...(((((..((((....((((.(((.....))).))))((((....)))).....))))..)))))(((((((....))...(((((((((((....)))))))))....)).))))). ( -38.00) >DroYak_CAF1 20901 120 - 1 CCAGUCAUAGCAUUCAAUCGCUGUGCAAGGAGCAGAGUGUGGAAUUCUCCAAACUAGAUGUUAUGGCCAUGGCGAACUCAACGGGAUUGGAGGAUCUCUCCUUCAGUAAUGCCACCAUGU ...(((((((((((....((((.(((.....))).))))((((....)))).....)))))))))))(((((.(..((.....))(((((((((....))))))))).....).))))). ( -40.60) >consensus CCAGUCACAGCAUUCAAUCGCUGUGCAAGGAGCAGAGUGUGGAGUUCUCCAAACUAGAUGUUAUGGCCAUAGCGAACUCGACAGGAUUGGAGGAUAUCUCCUUCAGUAAUGCCACCAUGU .........(((((...(((((((((....((((.(((.((((....)))).)))...))))...).))))))))..........(((((((((....))))))))))))))........ (-32.16 = -32.72 + 0.56)


| Location | 20,668,527 – 20,668,647 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -39.94 |
| Consensus MFE | -36.26 |
| Energy contribution | -37.46 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20668527 120 - 22407834 GAACGCUAUAGGAUGCAGCAUUCGACCGAGGGUUCUGUGUCCAGUCACAGCAUUCAAUCGCUGUGCAAGGAGCAGAGUGUUGAGUUCUCCAAACUAGAUGUUAUGGCCAUAGCGAACUCG ...((((((.((((((((.((((......)))).)))))))).((((.(((((((((.((((.(((.....))).)))))))((((.....)))).)))))).)))).))))))...... ( -41.40) >DroSec_CAF1 21375 120 - 1 GAACGCUAUAGGAUGCAGCACUCGGCCGAGGGUUCUGUGUCCAGUCACAGCAUUCAAUCCCUGUGCAAGGAGCAGAGUGUGGAGUUCUCCAAACUAGAUGUUAUGGCCAUAGCGAACUCG ...((((((.((((((((.((((......)))).)))))))).((((.((((((......((((.......))))(((.((((....)))).))).)))))).)))).))))))...... ( -42.60) >DroSim_CAF1 21701 120 - 1 GAACGCUAUAGGAUGCAGCACUCGACCGAGGGUUCUGUGUCCAGUCACAGCAUUCAAUCCCUGUGCAAGGAGCAGAGUGUGGAGUUCUCCAAACUAGAUGUUAUGGCCAUAGCGAACUCG ...((((((.((((((((.((((......)))).)))))))).((((.((((((......((((.......))))(((.((((....)))).))).)))))).)))).))))))...... ( -42.60) >DroEre_CAF1 21011 120 - 1 GAACGCUAUAGAAUGCAGCAUUCAGCCGAGGGUUCUGUGUCCAGUCAUAGCAUUCAAUCGCUGUGCAAGGAGCAGAGUGUGGAGUUCUCCAAACUAGAUGUCAUGGCCAUGGCGAACUCG ...((((((.(((((...))))).((((.((..(((((.(((...((((((........))))))...))))))))((.((((....)))).))......)).)))).))))))...... ( -36.90) >DroYak_CAF1 20941 120 - 1 GAACGCUAUAGAAUGCAACAGUCAGCCGAGGGUUCUGUGUCCAGUCAUAGCAUUCAAUCGCUGUGCAAGGAGCAGAGUGUGGAAUUCUCCAAACUAGAUGUUAUGGCCAUGGCGAACUCA ...((((((....((..((((..((((...))))))))...))(((((((((((....((((.(((.....))).))))((((....)))).....))))))))))).))))))...... ( -36.20) >consensus GAACGCUAUAGGAUGCAGCACUCGGCCGAGGGUUCUGUGUCCAGUCACAGCAUUCAAUCGCUGUGCAAGGAGCAGAGUGUGGAGUUCUCCAAACUAGAUGUUAUGGCCAUAGCGAACUCG ...((((((.((((((((.((((......)))).)))))))).((((.((((((....((((.(((.....))).))))((((....)))).....)))))).)))).))))))...... (-36.26 = -37.46 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:07 2006