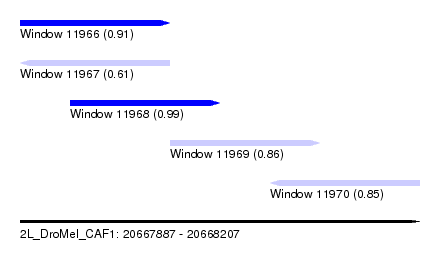
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,667,887 – 20,668,207 |
| Length | 320 |
| Max. P | 0.986063 |

| Location | 20,667,887 – 20,668,007 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20667887 120 + 22407834 UAACAAUUGUUUCUGAUUUUUUGCCUUGACUGGACUCGGUCAUGGUCUUCCCAUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUCGAGGUUCUCUUCACCUCCACCAGAGCUUCAU .............(((......(((.((((((....)))))).))).......(((((..((..(((((((..((.(((((......)))))))...)))))))..)))))))...))). ( -35.10) >DroSec_CAF1 20735 120 + 1 UAACAAUUGUUUCUGAUUUUUUGCCUUGACUGGACUCGGUCAUGGUCUUCCCAUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUCGAGGUUCUCUUCACCUCCACCAGAGCUUCAU .............(((......(((.((((((....)))))).))).......(((((..((..(((((((..((.(((((......)))))))...)))))))..)))))))...))). ( -35.10) >DroSim_CAF1 21061 120 + 1 UAACAAUUGUUUCUGAUUUUUUGCCUUGACUGGACUCGGUCAUGUUCUUCCCAUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUCGAGGUUCUCUUCACCUCCACCAGAGCUUCAU ......................((..((((((....))))))...........(((((..((..(((((((..((.(((((......)))))))...)))))))..)))))))))..... ( -31.50) >DroEre_CAF1 20371 120 + 1 UAACAAUUGUUUCCGGUUUUUUGCCUUGACUGGAUUCUGUCAUGGUUUUUCCCUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUGGAGGUUCUCUUCACUUCCACCAGAGCUUCAU ..............((......(((.((((........)))).)))....))((((((..(((.(((.(((.....))).)))))).(((((((.......)))))))))))))...... ( -35.40) >DroYak_CAF1 20301 120 + 1 UAGCUAUUGUUUCCGGUUUUUUACCUUGACUGGAUUCUGUCAUGGUUUUUCCAUCGGGAUUGCUGGUAAAGUUGCUCUUGACUGCAUUGGAGCUUCUCUUCACUUCUACCAGAGCUUCAU .((((.....(((((((.....(((.((((........)))).)))......)))))))...(((((((((((((........)))..((((.....)))))))).)))))))))).... ( -32.40) >consensus UAACAAUUGUUUCUGAUUUUUUGCCUUGACUGGACUCGGUCAUGGUCUUCCCAUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUCGAGGUUCUCUUCACCUCCACCAGAGCUUCAU ......................(((.((((((....)))))).))).......(((((..(((.(((.(((.....))).))))))...(((((.......)))))..)))))....... (-25.52 = -25.96 + 0.44)

| Location | 20,667,887 – 20,668,007 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -23.50 |
| Energy contribution | -22.86 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20667887 120 - 22407834 AUGAAGCUCUGGUGGAGGUGAAGAGAACCUCGAAUGCGGUCAAGAGCAACUUCACCAGCAAUCCAGAUGGGAAGACCAUGACCGAGUCCAGUCAAGGCAAAAAAUCAGAAACAAUUGUUA .....((..((((((((((.......)))))((...((((((.(.(...((((.(((.(......).))))))).)).))))))..)))).)))..))...................... ( -29.70) >DroSec_CAF1 20735 120 - 1 AUGAAGCUCUGGUGGAGGUGAAGAGAACCUCGAAUGCGGUCAAGAGCAACUUCACCAGCAAUCCAGAUGGGAAGACCAUGACCGAGUCCAGUCAAGGCAAAAAAUCAGAAACAAUUGUUA .....((..((((((((((.......)))))((...((((((.(.(...((((.(((.(......).))))))).)).))))))..)))).)))..))...................... ( -29.70) >DroSim_CAF1 21061 120 - 1 AUGAAGCUCUGGUGGAGGUGAAGAGAACCUCGAAUGCGGUCAAGAGCAACUUCACCAGCAAUCCAGAUGGGAAGAACAUGACCGAGUCCAGUCAAGGCAAAAAAUCAGAAACAAUUGUUA .....((..((((((((((.......)))))((...((((((.(.....((((.(((.(......).)))))))..).))))))..)))).)))..))...................... ( -29.20) >DroEre_CAF1 20371 120 - 1 AUGAAGCUCUGGUGGAAGUGAAGAGAACCUCCAAUGCGGUCAAGAGCAACUUCACCAGCAAUCCAGAGGGAAAAACCAUGACAGAAUCCAGUCAAGGCAAAAAACCGGAAACAAUUGUUA ......((((((((((.((.......)).)))).((((((.(((.....))).))).)))..))))))((.....((.((((........)))).)).......))(....)........ ( -29.40) >DroYak_CAF1 20301 120 - 1 AUGAAGCUCUGGUAGAAGUGAAGAGAAGCUCCAAUGCAGUCAAGAGCAACUUUACCAGCAAUCCCGAUGGAAAAACCAUGACAGAAUCCAGUCAAGGUAAAAAACCGGAAACAAUAGCUA ....((((((((((((..((....((.((......))..)).....))..))))))))...(((....)))...(((.((((........)))).)))........(....)...)))). ( -25.60) >consensus AUGAAGCUCUGGUGGAGGUGAAGAGAACCUCGAAUGCGGUCAAGAGCAACUUCACCAGCAAUCCAGAUGGGAAGACCAUGACCGAGUCCAGUCAAGGCAAAAAAUCAGAAACAAUUGUUA .......((((((...(((((((.(.....)...(((........))).))))))).((..(((....))).......((((........))))..)).....))))))........... (-23.50 = -22.86 + -0.64)


| Location | 20,667,927 – 20,668,047 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -30.46 |
| Energy contribution | -30.54 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20667927 120 + 22407834 CAUGGUCUUCCCAUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUCGAGGUUCUCUUCACCUCCACCAGAGCUUCAUAUUUCUCGACUAGGUCCCGAUAAGGAUUUGGUUUGUCGCU ..(((.....)))(((((..((..(((((((..((.(((((......)))))))...)))))))..)))))))((..((........((((((((((......))))))))))))..)). ( -36.80) >DroSec_CAF1 20775 120 + 1 CAUGGUCUUCCCAUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUCGAGGUUCUCUUCACCUCCACCAGAGCUUCAUAUUUCUCGACUAGGUCCCGAUAGGGAUUUGGUUUGUCGCU ..(((.....)))(((((..((..(((((((..((.(((((......)))))))...)))))))..)))))))((..((........(((((((((((....)))))))))))))..)). ( -41.10) >DroSim_CAF1 21101 120 + 1 CAUGUUCUUCCCAUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUCGAGGUUCUCUUCACCUCCACCAGAGCUUCAUAUUUCUCGACUAGGUCCCGAUAGGGAUUUGGUUUGUCGCU .............(((((..((..(((((((..((.(((((......)))))))...)))))))..)))))))((..((........(((((((((((....)))))))))))))..)). ( -39.90) >DroEre_CAF1 20411 120 + 1 CAUGGUUUUUCCCUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUGGAGGUUCUCUUCACUUCCACCAGAGCUUCAUACUUCUCGACUAGAUCCCGAUAGGGAUUUGGUUUCUCGCC ...(((......((((((..(((.(((.(((.....))).)))))).(((((((.......))))))))))))).............(((((((((((....)))))))))))....))) ( -42.20) >DroYak_CAF1 20341 120 + 1 CAUGGUUUUUCCAUCGGGAUUGCUGGUAAAGUUGCUCUUGACUGCAUUGGAGCUUCUCUUCACUUCUACCAGAGCUUCAUAUUUCUCGACUAGGUCUCGAUACGGAUUUGGUUUGUCGCU .((((.....))))(((((.(((.(((.(((.....))).)))))).((((((((................))))))))....)))))(((((((((......)))))))))........ ( -26.79) >consensus CAUGGUCUUCCCAUCUGGAUUGCUGGUGAAGUUGCUCUUGACCGCAUUCGAGGUUCUCUUCACCUCCACCAGAGCUUCAUAUUUCUCGACUAGGUCCCGAUAGGGAUUUGGUUUGUCGCU .............(((((..(((.(((.(((.....))).))))))...(((((.......)))))..)))))..............(((((((((((....)))))))))))....... (-30.46 = -30.54 + 0.08)

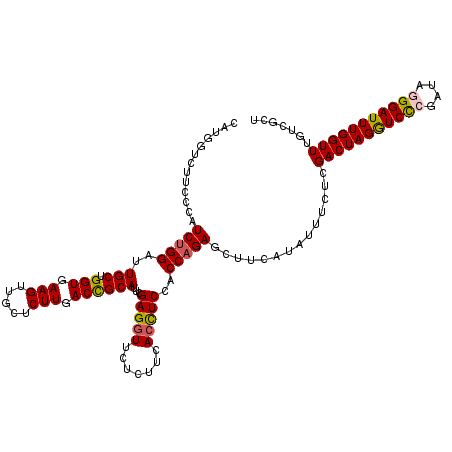

| Location | 20,668,007 – 20,668,127 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -34.30 |
| Energy contribution | -33.82 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20668007 120 + 22407834 AUUUCUCGACUAGGUCCCGAUAAGGAUUUGGUUUGUCGCUAGAGUUGGGAUUGAGGUCCUCGGGAAUCCGGAAGCGUUCCUUAUAAUCCGCACCACUAUUAUCGCAUUUCGAGGUGCUUU .......((((.(((((((((.....((((((.....)))))))))))))))..))))...((((((.(....).))))))........(((((.......(((.....))))))))... ( -32.91) >DroSec_CAF1 20855 120 + 1 AUUUCUCGACUAGGUCCCGAUAGGGAUUUGGUUUGUCGCUAGAGUUGGGAUUGAGGUCCUCGGGAAUCCGGAAGCGUUCCUUAUAAUCCGCAGCACUAUUAUCGCAUUUCGAGGUGCUUU ...(((((((((((((((....)))))))))......(((((.((((((((((((...)))((((((.(....).))))))...))))).)))).))).....))...))))))...... ( -40.10) >DroSim_CAF1 21181 120 + 1 AUUUCUCGACUAGGUCCCGAUAGGGAUUUGGUUUGUCGCUAGAGUUGGGAUUGAGGUCCUCGGGAAUCCGGAAGCGUUCCUUAUAAUCCGCAGCACUAUUAUCGCAUUUCGAGGUGCUUU ...(((((((((((((((....)))))))))......(((((.((((((((((((...)))((((((.(....).))))))...))))).)))).))).....))...))))))...... ( -40.10) >DroEre_CAF1 20491 120 + 1 ACUUCUCGACUAGAUCCCGAUAGGGAUUUGGUUUCUCGCCAGAGUUGGGAUUAAGGUCCUCGGAAAUCCGAAAGCGUUCCUUAUAAUCCGCAGCACUGUUAUCGCACUUCGAGGUGCUUU .......(((((((((((....)))))))))))......(((.(((((((((((((..(((((....))))....)..)))..)))))).)))).))).....(((((....)))))... ( -42.60) >DroYak_CAF1 20421 120 + 1 AUUUCUCGACUAGGUCUCGAUACGGAUUUGGUUUGUCGCUAGAGUUCGGAUUAAGGUCCUCGGGAAUCCGAAAGCGUUCCUUAUAGUCCGCAGCACUAUUAUCACAUUUCGAGGUGCUUU .......((((((..((..((.((((((((((.....)))))).)))).))..))..)...((((((.(....).))))))..)))))...((((((...............)))))).. ( -31.16) >consensus AUUUCUCGACUAGGUCCCGAUAGGGAUUUGGUUUGUCGCUAGAGUUGGGAUUGAGGUCCUCGGGAAUCCGGAAGCGUUCCUUAUAAUCCGCAGCACUAUUAUCGCAUUUCGAGGUGCUUU ...(((((((((((((((....)))))))))........(((.(((((((((((((..(((((....))))....)..)))))..)))).)))).)))..........))))))...... (-34.30 = -33.82 + -0.48)

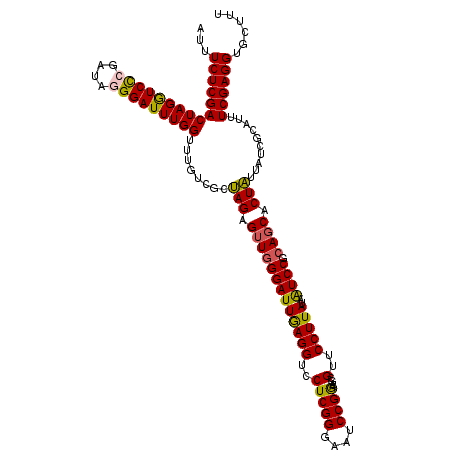
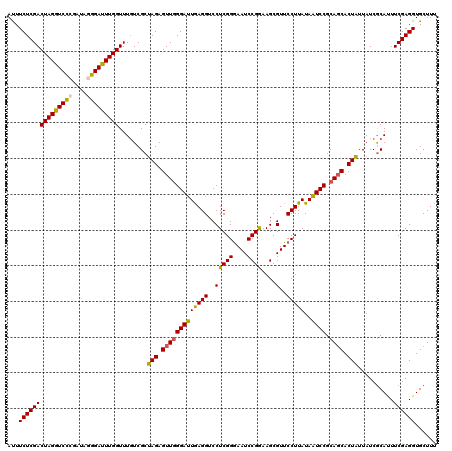
| Location | 20,668,087 – 20,668,207 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -26.99 |
| Consensus MFE | -25.60 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20668087 120 - 22407834 CAACACCAAUAUGGAUGAUCAAUCCUCCAUUGCCCCAACCGAAGAAAUCUACGAAGAUGACGAUCGCAGCAUACUAAGCGAAAGCACCUCGAAAUGCGAUAAUAGUGGUGCGGAUUAUAA .....((...(((((.((....)).))))).((.(((..((.((....)).)).........((((((.........((....)).........)))))).....))).))))....... ( -26.47) >DroSec_CAF1 20935 120 - 1 CAACACCAAUAUGGAUGAUCAAUCCUCCAUUGCGCCAACCGAAGAAAUCUACGAAGAUGACGAUCGCAGCAUACUAAGCGAAAGCACCUCGAAAUGCGAUAAUAGUGCUGCGGAUUAUAA ......((((..((((.....))))...))))......((((.(..((((....))))..)..))(((((((.....((....))...(((.....))).....)))))))))....... ( -27.40) >DroSim_CAF1 21261 120 - 1 CAACACCAAUAUGGAUGAUCAAUCCUCCAUUGCGCCAACCGAAGAAAUCUACGAAGAUGACGAUCGCAGCAUACUAAGCGAAAGCACCUCGAAAUGCGAUAAUAGUGCUGCGGAUUAUAA ......((((..((((.....))))...))))......((((.(..((((....))))..)..))(((((((.....((....))...(((.....))).....)))))))))....... ( -27.40) >DroEre_CAF1 20571 120 - 1 UAACACCAAUAUGGAUGAUCAGUCCUCCAUUGCUCCAACCGAAGAAAUCUAUGAAGAUGACGAUCGCAGCAUACUAAGCGAAAGCACCUCGAAGUGCGAUAACAGUGCUGCGGAUUAUAA ..........(((((.((....)).)))))...(((....((.(..((((....))))..)..))(((((((.....((....))...(((.....))).....))))))))))...... ( -28.30) >DroYak_CAF1 20501 120 - 1 UAAUACCAAUAUGGAUGAUCAAUCCUCCAUUGCUCCAACCGAAGAAAUUUAUGAAGAUGACGAUCGCAGCAUAUUAAGCGAAAGCACCUCGAAAUGUGAUAAUAGUGCUGCGGACUAUAA ......((((..((((.....))))...)))).(((...................(((....)))(((((((((((.((....))...(((.....))))))).))))))))))...... ( -25.40) >consensus CAACACCAAUAUGGAUGAUCAAUCCUCCAUUGCGCCAACCGAAGAAAUCUACGAAGAUGACGAUCGCAGCAUACUAAGCGAAAGCACCUCGAAAUGCGAUAAUAGUGCUGCGGAUUAUAA ......((((..((((.....))))...))))......((((.(..((((....))))..)..))(((((((.....((....))...(((.....))).....)))))))))....... (-25.60 = -25.32 + -0.28)


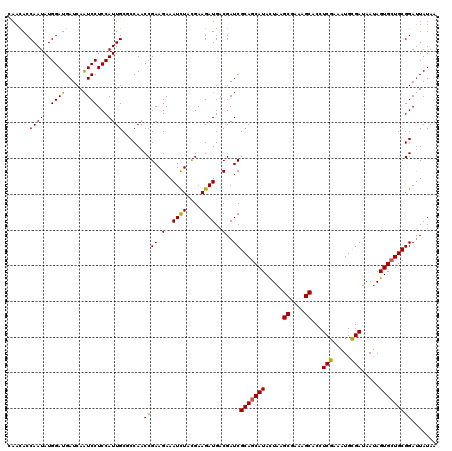
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:04 2006