
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,652,648 – 20,652,777 |
| Length | 129 |
| Max. P | 0.939853 |

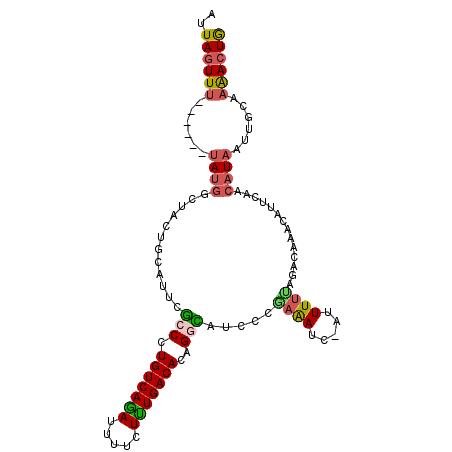
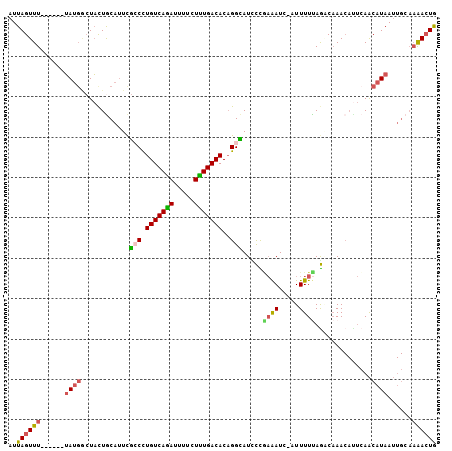
| Location | 20,652,648 – 20,652,747 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -20.63 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.52 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20652648 99 - 22407834 AUUAGUUU------UAUGGCUACUACAUUCGCCCUGUCAGAUUUUCUUUGACACAGGCAUCCCGAAAUC-AUUUUUAGCCAAACAUUCAACAUAAUUGCAAAACUG ..((((((------(.((((((........(((.(((((((.....)))))))..))).....(....)-.....)))))).........((....)).))))))) ( -20.10) >DroSec_CAF1 5452 99 - 1 AUUAGUUU------UAUGACUACUGCAUUCGCCCUGUCAGAUUUUCUUUGACACAGGCAUCCCAAAAUC-AUUUUUAGGCAAACAUUCAACAUAAUUGCAAAACUG ..(((((.------...))))).((((...(((.(((((((.....)))))))..)))..((.((((..-..)))).)).................))))...... ( -18.00) >DroSim_CAF1 5455 99 - 1 AUUAGUUU------UAUGGCUACUGCAUUCGCCCUGUCAGAUUUUCUUUGACACAGGCAUCCCGAAAUC-AUUUUUAGCCAAACAUUCAACAUAAUUGCAAAACUG ..((((((------(.((((((........(((.(((((((.....)))))))..))).....(....)-.....)))))).........((....)).))))))) ( -20.10) >DroEre_CAF1 5489 99 - 1 AUUAGUUU------UAUGGCUACUGCACUCGCCCUGUCAGAUUUUCUCUGACACAGGUAUUCUGAAAUC-AUUUUCAGACAAACACUCAACAUAAUUGCAAAACUG ....((.(------((((((....))....(((.(((((((.....)))))))..)))..(((((((..-..)))))))...........)))))..))....... ( -23.20) >DroYak_CAF1 5507 105 - 1 AUUAGUUUGCUUAUUAGUUAUACUGCAUUCAGCCUGUCAAAUUUUCUUUGACACAGGUAUUUUAAAAUC-UUUUUUAAGCAAACAUUCAACAUAAUUGCAAAACUG ....(((((((((...((......)).....((((((((((.....))))))..))))...........-.....)))))))))...................... ( -19.50) >DroAna_CAF1 5417 90 - 1 AUUAGUUU------GAUGACUUUUGCAUUUGGCUUGUCAGAAAUCCUCUGACACUGGCAUCUUUCGAUCCAUUUUCAAACCAA----AAGCAUAA------GAAUA ........------.....(((.(((.(((((..(((((((.....))))))).(((.(((....))))))........))))----).))).))------).... ( -22.90) >consensus AUUAGUUU______UAUGGCUACUGCAUUCGCCCUGUCAGAUUUUCUUUGACACAGGCAUCCCGAAAUC_AUUUUUAGACAAACAUUCAACAUAAUUGCAAAACUG ..((((((......((((............(((.(((((((.....)))))))..))).....((((.....))))..............))))......)))))) (-11.43 = -11.52 + 0.09)

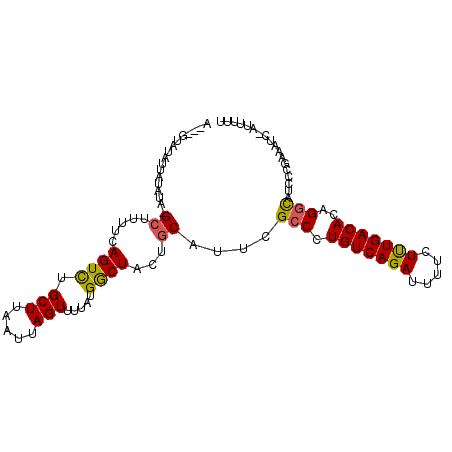
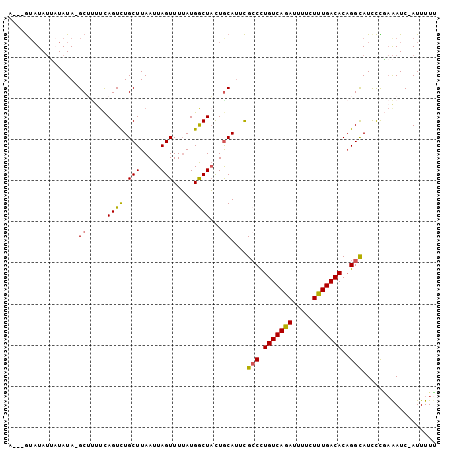
| Location | 20,652,678 – 20,652,777 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -19.04 |
| Consensus MFE | -14.18 |
| Energy contribution | -13.78 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20652678 99 - 22407834 A---GUAUAUUAUAUA-GCUUUUCAGUUUGCUUAAUUAGUUUUAUGGCUACUACAUUCGCCCUGUCAGAUUUUCUUUGACACAGGCAUCCCGAAAUC-AUUUUU .---............-...((((.(..(((((............(((..........))).(((((((.....))))))).)))))..).))))..-...... ( -16.30) >DroSec_CAF1 5482 99 - 1 A---GUAUAUUAUAUA-GCUUUUCAGUCUGCUUAAUUAGUUUUAUGACUACUGCAUUCGCCCUGUCAGAUUUUCUUUGACACAGGCAUCCCAAAAUC-AUUUUU (---(((..(((((.(-(((.((.((....)).))..)))).))))).))))......(((.(((((((.....)))))))..)))...........-...... ( -16.50) >DroSim_CAF1 5485 99 - 1 A---GUAUAUUAUAUA-GCUUUUCAGUCUGCUUAAUUAGUUUUAUGGCUACUGCAUUCGCCCUGUCAGAUUUUCUUUGACACAGGCAUCCCGAAAUC-AUUUUU .---............-...((((.(..(((.....(((((....)))))..)))..)(((.(((((((.....)))))))..))).....))))..-...... ( -17.50) >DroEre_CAF1 5519 93 - 1 A---G--UAUACUAUA-GC----CAGUCUGCUUAAUUAGUUUUAUGGCUACUGCACUCGCCCUGUCAGAUUUUCUCUGACACAGGUAUUCUGAAAUC-AUUUUC (---(--.(((((.((-((----((....(((.....)))....))))))............(((((((.....)))))))..))))).))......-...... ( -22.30) >DroAna_CAF1 5437 104 - 1 AAUAAUAUAAACUAUAUGCACUUUAGUCUGCUUAAUUAGUUUGAUGACUUUUGCAUUUGGCUUGUCAGAAAUCCUCUGACACUGGCAUCUUUCGAUCCAUUUUC ...........(((.(((((....((((.(((.....))).....))))..))))).)))..(((((((.....))))))).(((.(((....))))))..... ( -22.60) >consensus A___GUAUAUUAUAUA_GCUUUUCAGUCUGCUUAAUUAGUUUUAUGGCUACUGCAUUCGCCCUGUCAGAUUUUCUUUGACACAGGCAUCCCGAAAUC_AUUUUU .................((.....((((.(((.....))).....))))...))....(((.(((((((.....)))))))..))).................. (-14.18 = -13.78 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:57 2006