
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,648,349 – 20,648,455 |
| Length | 106 |
| Max. P | 0.604155 |

| Location | 20,648,349 – 20,648,455 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -20.98 |
| Energy contribution | -22.07 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
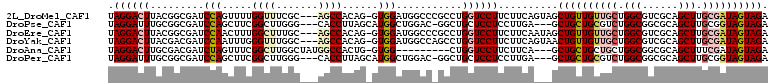
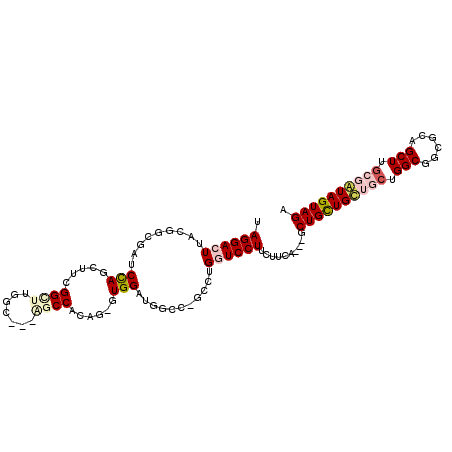
>2L_DroMel_CAF1 20648349 106 + 22407834 UAGGACUUACGGCGAUCCAGUUUUGGUUUCGC---AGCCACAG-GUGGAUGGCCCGCCUGGUCCUUCUUCAGUAGCUGUUGUUGCUGGCGUCGCAGCUUGCGAUAGUAGA .((((((...(((.(((((.((.((((.....---.)))).))-.))))).))).....))))))...((((((((....)))))))).(((((.....)))))...... ( -42.80) >DroPse_CAF1 949 103 + 1 UAGGAUUUGCGGCGAUCCAGCUUCGGCUUGGG---CACCUUAGCAUGGCUGGAC-GGCUGCUCCUCCUUGA---GCUGCUGCGUCUGGCGGCGCAGCUUGCGGUAGUAGA .((((...(((((..(((((((...(((.((.---...)).)))..))))))).-.)))))...))))...---((.((((((((....))))))))..))......... ( -49.10) >DroEre_CAF1 1133 106 + 1 UAGGACUUACGGCGAUCCAACUUUGGCUUUGC---AGCCACAG-GUGGAUGGCCCGCCUGGUCCUUCUUCAAUAGCUGUUGUUGCUGGCGUCGCAGCUUGCGAUAGUAGA .((((((...(((.(((((.((.(((((....---))))).))-.))))).))).....)))))).........((((((((.((((......))))..))))))))... ( -44.20) >DroYak_CAF1 1362 106 + 1 UAGGACUUACGACGAUCCAAUUUGGGUUUGGC---AGCCACAG-GUGGAUGGCCAGCCUGGUCCUUCUUCAGUAACUGUUGUUGCUGGCGUCGCAGCUUGCGAUAGUAGA .((((((......(((((.....)))))((((---..(((...-.)))...))))....))))))...((((((((....)))))))).(((((.....)))))...... ( -36.40) >DroAna_CAF1 1122 97 + 1 UAGGACUUGCGACGAUCUAGUUUCGGCUUGGCUAUGGCCACUG-GUGG---------CUGGUCCUUCUUCA---GCUGCUGCUGCUGGCGGCGCAGCUUUCGAUAGUAGA .((((((.((.((((.......))((((.......))))....-)).)---------).))))))((...(---(((((.((((....))))))))))...))....... ( -35.20) >DroPer_CAF1 949 103 + 1 UAGGAUUUGCGGCGAUCCAGCUUCGGCUUGGG---CACCUUAGCAUGGCUGGAC-GGCUGCUCCUCCUUGA---GCUGCUGCGUCUGGCGGCGCAGCUUGCGGUAGUAGA .((((...(((((..(((((((...(((.((.---...)).)))..))))))).-.)))))...))))...---((.((((((((....))))))))..))......... ( -49.10) >consensus UAGGACUUACGGCGAUCCAGCUUCGGCUUGGC___AGCCACAG_GUGGAUGGCC_GCCUGGUCCUUCUUCA___GCUGCUGCUGCUGGCGGCGCAGCUUGCGAUAGUAGA .((((((.........(((.....((((.......))))......)))...........))))))..........((((((((((.(((......))).)))))))))). (-20.98 = -22.07 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:54 2006