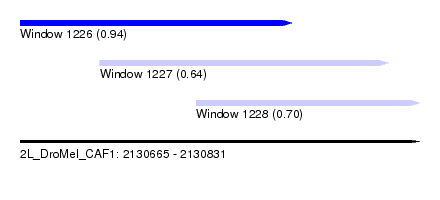
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,130,665 – 2,130,831 |
| Length | 166 |
| Max. P | 0.936511 |

| Location | 2,130,665 – 2,130,778 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -34.75 |
| Energy contribution | -36.87 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2130665 113 + 22407834 UGUUGUAUUAUAUCCCCUAGAUGAUAAU-------UCGUAGCAGUUGAUACCCAGCGCACCCACUGUGCGCUGGGAGUUGUUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCG ((((((......((((((....((((((-------(((((........)))(((((((((.....)))))))))))))))))....))))))))))))((((.((((.....)))))))) ( -47.70) >DroSim_CAF1 130061 120 + 1 UAUUGAAUUGUAUCCCCUUGAUGAUAAUUAUCUGUUCAUAACAGUUGAUACUCAGCGCACCCACUGUGCGCUGGGAGUUGUUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCG .......((((.((((((((..(((((((..((((.....))))......((((((((((.....)))))))))))))))))..))))))))))))..((((.((((.....)))))))) ( -47.80) >DroYak_CAF1 136131 116 + 1 UCUUGCAUUUUAUCCCUUUGAUGAUAAGUAUUUGUUCGUAGCAGUUGAGACACAGC----CCACUGUGCGCUGGGAGUUGUUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCG ...(((......((((((((..(((((.......(((.((((......(.(((((.----...)))))))))).))))))))..))))))))...)))((((.((((.....)))))))) ( -31.21) >consensus UAUUGAAUUAUAUCCCCUUGAUGAUAAUUAU_UGUUCGUAGCAGUUGAUACACAGCGCACCCACUGUGCGCUGGGAGUUGUUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCG ............((((((((..(((((((......(((.......)))..((((((((((.....)))))))))))))))))..))))))))......((((.((((.....)))))))) (-34.75 = -36.87 + 2.12)

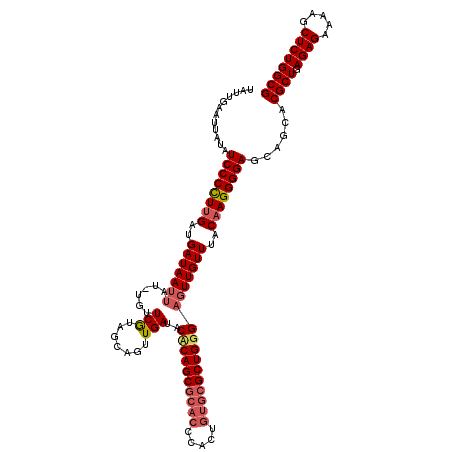
| Location | 2,130,698 – 2,130,818 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -32.58 |
| Energy contribution | -34.47 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2130698 120 + 22407834 GCAGUUGAUACCCAGCGCACCCACUGUGCGCUGGGAGUUGUUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCGCAAAAGCUAUCUCCGAAAGUAUGCCGAAAGUAUAAAGUGA ......(((.((((((((((.....)))))))))).)))..((((..(((((..(((.((((.((((.....)))))))).....))).))))).....((((.(....)))))..)))) ( -43.20) >DroSim_CAF1 130101 120 + 1 ACAGUUGAUACUCAGCGCACCCACUGUGCGCUGGGAGUUGUUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCGCAAAAGCUAUCUCCGAAAGUAUUCCGAGAGUAUAAAGUGA .((.((.(((((((((((((.....))))))).(((((..(((....(((((..(((.((((.((((.....)))))))).....))).))))).)))..)))))..)))))).)).)). ( -44.30) >DroYak_CAF1 136171 116 + 1 GCAGUUGAGACACAGC----CCACUGUGCGCUGGGAGUUGUUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCGCAAAAGCUAUCUCCGAAAGUAUGGCGAAAGUAUAAAGUGA ...((((.....))))----.((((.(((((..((.(((((((.......)))))))..(((........)))))..)))))...(((((...(....).)))))..........)))). ( -33.40) >consensus GCAGUUGAUACACAGCGCACCCACUGUGCGCUGGGAGUUGUUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCGCAAAAGCUAUCUCCGAAAGUAUGCCGAAAGUAUAAAGUGA ..........((((((((((.....))))))))))......((((..(((((..(((.((((.((((.....)))))))).....))).))))).....((((.(....)))))..)))) (-32.58 = -34.47 + 1.89)

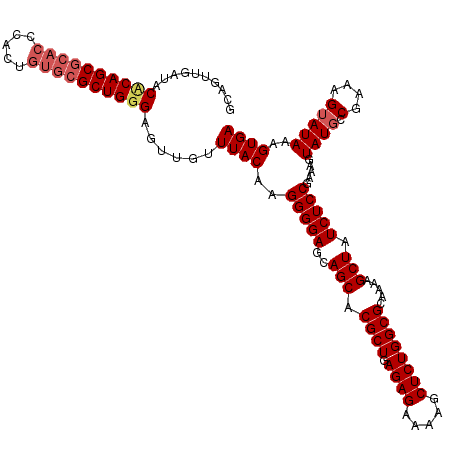
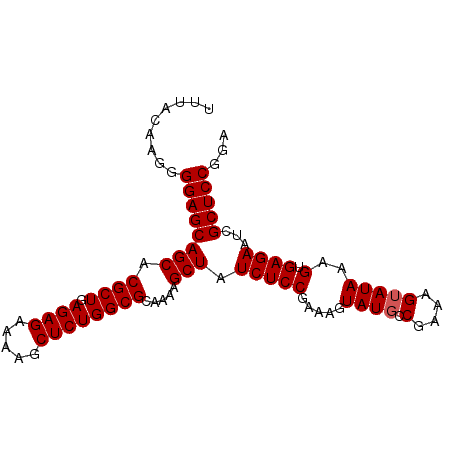
| Location | 2,130,738 – 2,130,831 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 97.85 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2130738 93 + 22407834 UUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCGCAAAAGCUAUCUCCGAAAGUAUGCCGAAAGUAUAAAGUGAGAAUCGCUCCGGA .........((((((((.((((.((((.....)))))))).....))).(((((.....((((.(....)))))..).))))...)))))... ( -26.20) >DroSim_CAF1 130141 93 + 1 UUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCGCAAAAGCUAUCUCCGAAAGUAUUCCGAGAGUAUAAAGUGAGAAUCGCUCCGGA .........((((((((.((((.((((.....)))))))).....))).(((((....((((((...))))))...).))))...)))))... ( -25.80) >DroYak_CAF1 136207 93 + 1 UUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCGCAAAAGCUAUCUCCGAAAGUAUGGCGAAAGUAUAAAGUGAGAAUCGCUCCGGA .........((((((((.((((.((((.....)))))))).....))).(((((....)....((....)).......))))...)))))... ( -26.60) >consensus UUUACAAGGGGAGCAGCACGCUGAGAGAAAAGCUCUGGCGCAAAAGCUAUCUCCGAAAGUAUGCCGAAAGUAUAAAGUGAGAAUCGCUCCGGA .........((((((((.((((.((((.....)))))))).....))).(((((.....((((.(....)))))..).))))...)))))... (-24.37 = -24.70 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:36 2006