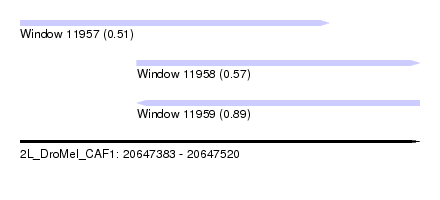
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,647,383 – 20,647,520 |
| Length | 137 |
| Max. P | 0.888548 |

| Location | 20,647,383 – 20,647,489 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
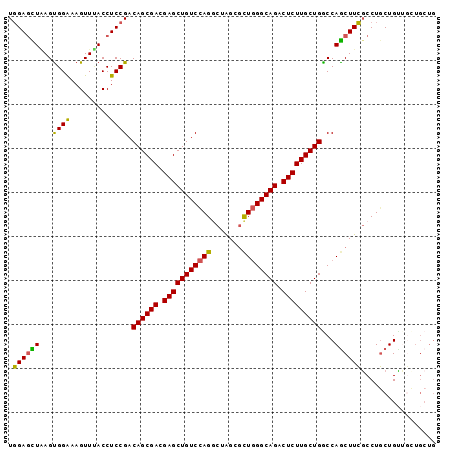
>2L_DroMel_CAF1 20647383 106 + 22407834 UCGCCCAGUAUGCGAAACGAAUUCUGUGUGGUGGCGGUGCUGGAGCUAAGUGGGAAGUUUACCUCCGACAGCGACGAGCUGUCCAGACUGGCGCUGGGCA-----GACUCU ((((.......)))).......(((((.(((((.((((.(((((.....((.(((.(....).))).))(((.....))).))))))))).))))).)))-----)).... ( -39.60) >DroGri_CAF1 131 99 + 1 UCGCCAAGUAUGCGAAAUGAGUUUUGCGU------------CGAAUUAAACGGAAAAUUUAUCUCCGACAGGGAUGAGCUGUCCAGACUGGCAUUGGGUGUCAGCGAUACG ..((((.((((((((((....))))))))------------..........(((...((((((.((....))))))))...)))..)))))).....(((((...))))). ( -27.00) >DroSec_CAF1 179 100 + 1 UCGCCCAGUAUGCGAAACGAAUUCUGUG------CGGUGCUGGAGUUAAGUGGAAAGUUUACCUCCGACAGCGACGAGCUGUCCAGGCUAGCGCUGGGCA-----GACUCU ((((.......)))).......(((((.------(((((((((((.(((((....).)))).))))((((((.....))))))......))))))).)))-----)).... ( -38.40) >DroSim_CAF1 179 100 + 1 UCGCCCAGUAUGCGAAACGAAUUCUGUG------CGGUGCUGGAGUUAAGUGGAAAGUUUACCUCCGACAGCGACGAGCUGUCCAGGCUAGCGCUGGGCA-----GACUCU ((((.......)))).......(((((.------(((((((((((.(((((....).)))).))))((((((.....))))))......))))))).)))-----)).... ( -38.40) >DroYak_CAF1 399 103 + 1 UCGCCCAGUAUCCGAAAGGAAUUCUGUGU---GACGGUGCUGGAGCUAAGUGGAAAGUUCACCUCCGACAGCGACGAGCUGUCCAGGCUAGGGCUGGGCA-----GACUCU ((((.(((..(((....)))...))).))---))((.((.(((((....(((((...)))))))))).)).))..(((((((((((.(...).)))))))-----).))). ( -43.20) >DroAna_CAF1 173 106 + 1 UCGCCGAGUAUCCGAAACGAGUUCUGGGUGGCCUCAUUCGUCGACUUGAGCGGGAAGUUCACCUCCGACAGCGAGGAGCUGUCCAGACUGGCAUUGGGCA-----GGCUCU .....((((.(((((..(.(((.(((((..(((((.((.((((...(((((.....)))))....)))))).)))..))..)))))))).)..)))))..-----.)))). ( -37.80) >consensus UCGCCCAGUAUGCGAAACGAAUUCUGUGU_____CGGUGCUGGAGUUAAGUGGAAAGUUUACCUCCGACAGCGACGAGCUGUCCAGACUAGCGCUGGGCA_____GACUCU ..(((((((..((............................((((.(((((.....))))).))))((((((.....)))))).......)))))))))............ (-24.22 = -24.62 + 0.39)



| Location | 20,647,423 – 20,647,520 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 87.01 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -32.51 |
| Energy contribution | -31.68 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20647423 97 + 22407834 UGGAGCUAAGUGGGAAGUUUACCUCCGACAGCGACGAGCUGUCCAGACUGGCGCUGGGCAGACUCUUGCUGACCAGCUUCGCCUGCUGUUGCUGCUG .((((((..((.(((.(....).))).))(((((.(((((((((((.......)))))))).))))))))....))))))((..((....)).)).. ( -39.60) >DroSec_CAF1 213 97 + 1 UGGAGUUAAGUGGAAAGUUUACCUCCGACAGCGACGAGCUGUCCAGGCUAGCGCUGGGCAGACUCUUGCUGGCCAACUUCGCCUGCUGUUGCUGCUG .((((.(((((....).)))).))))(.(((((((((((((((((((....).)))))))).)))..((.(((.......))).)).)))))))).. ( -40.30) >DroSim_CAF1 213 97 + 1 UGGAGUUAAGUGGAAAGUUUACCUCCGACAGCGACGAGCUGUCCAGGCUAGCGCUGGGCAGACUCUUGCUGGCCAGCUUCGCCUGCUGUUGCUGCUG .((((.(((((....).)))).))))(.(((((((((((((((((((....).)))))))).)))..((.(((.......))).)).)))))))).. ( -40.30) >DroEre_CAF1 216 89 + 1 UGGAGCUCAGUGGAAAGUUGACCUCCGACAGCGACGAGCUGUCUAGGCUAGCGCUUGGCAGACUCUUGCUGGCCAGCUUCGCCUGCUGC-------- .((((.(((((....).)))).))))..((((((.((((((.((((((....))))))))).)))))))))..((((.......)))).-------- ( -36.10) >DroYak_CAF1 436 97 + 1 UGGAGCUAAGUGGAAAGUUCACCUCCGACAGCGACGAGCUGUCCAGGCUAGGGCUGGGCAGACUCUUGCUGGCCAGCUUCGCCUGCUGUUGUUGCUG .((((....(((((...)))))))))..(((((((.(((....(((((..(((((((.(((.(....)))).))))))).)))))..)))))))))) ( -42.30) >DroAna_CAF1 213 97 + 1 UCGACUUGAGCGGGAAGUUCACCUCCGACAGCGAGGAGCUGUCCAGACUGGCAUUGGGCAGGCUCUUGCUGGCCAGCUUGUCCUUCUGGAUCUGCUG (((...(((((.....)))))....)))(((((((.((((((((((.......))))))).))))))))))..((((..((((....))))..)))) ( -38.70) >consensus UGGAGCUAAGUGGAAAGUUUACCUCCGACAGCGACGAGCUGUCCAGGCUAGCGCUGGGCAGACUCUUGCUGGCCAGCUUCGCCUGCUGUUGCUGCUG .((((((...((((.........)))).((((((.(((((((((((.......)))))))).)))))))))...))))))................. (-32.51 = -31.68 + -0.83)

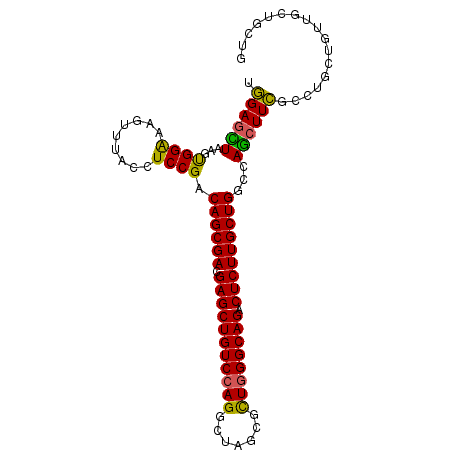
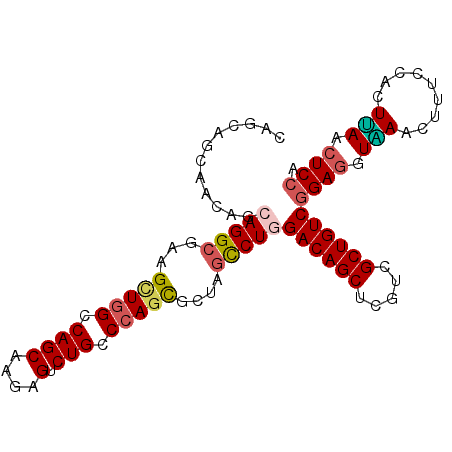
| Location | 20,647,423 – 20,647,520 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.01 |
| Mean single sequence MFE | -36.11 |
| Consensus MFE | -31.58 |
| Energy contribution | -31.72 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20647423 97 - 22407834 CAGCAGCAACAGCAGGCGAAGCUGGUCAGCAAGAGUCUGCCCAGCGCCAGUCUGGACAGCUCGUCGCUGUCGGAGGUAAACUUCCCACUUAGCUCCA ..(.(((.....(((((...(((((.((((....).))).)))))....)))))((((((.....))))))(((((....)))))......))).). ( -38.10) >DroSec_CAF1 213 97 - 1 CAGCAGCAACAGCAGGCGAAGUUGGCCAGCAAGAGUCUGCCCAGCGCUAGCCUGGACAGCUCGUCGCUGUCGGAGGUAAACUUUCCACUUAACUCCA ............(((((...(((((.((((....).))).)))))....)))))((((((.....))))))((((.................)))). ( -34.53) >DroSim_CAF1 213 97 - 1 CAGCAGCAACAGCAGGCGAAGCUGGCCAGCAAGAGUCUGCCCAGCGCUAGCCUGGACAGCUCGUCGCUGUCGGAGGUAAACUUUCCACUUAACUCCA ............(((((...(((((.((((....).))).)))))....)))))((((((.....))))))((((.................)))). ( -36.93) >DroEre_CAF1 216 89 - 1 --------GCAGCAGGCGAAGCUGGCCAGCAAGAGUCUGCCAAGCGCUAGCCUAGACAGCUCGUCGCUGUCGGAGGUCAACUUUCCACUGAGCUCCA --------.....((((...((((((..((....))..))).)))....)))).((((((.....))))))((((.(((.........))).)))). ( -35.40) >DroYak_CAF1 436 97 - 1 CAGCAACAACAGCAGGCGAAGCUGGCCAGCAAGAGUCUGCCCAGCCCUAGCCUGGACAGCUCGUCGCUGUCGGAGGUGAACUUUCCACUUAGCUCCA ............(((((...(((((.((((....).))).)))))....)))))((((((.....))))))((((.(((.........))).)))). ( -37.90) >DroAna_CAF1 213 97 - 1 CAGCAGAUCCAGAAGGACAAGCUGGCCAGCAAGAGCCUGCCCAAUGCCAGUCUGGACAGCUCCUCGCUGUCGGAGGUGAACUUCCCGCUCAAGUCGA .(((...(((....)))((..(((((..(((......))).....)))))..))((((((.....))))))(((((....))))).)))........ ( -33.80) >consensus CAGCAGCAACAGCAGGCGAAGCUGGCCAGCAAGAGUCUGCCCAGCGCUAGCCUGGACAGCUCGUCGCUGUCGGAGGUAAACUUUCCACUUAACUCCA ............(((((...(((((.((((....).))).)))))....)))))((((((.....))))))((((.(((.........))).)))). (-31.58 = -31.72 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:53 2006