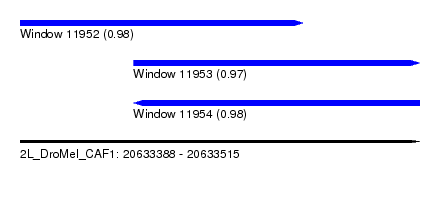
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,633,388 – 20,633,515 |
| Length | 127 |
| Max. P | 0.981559 |

| Location | 20,633,388 – 20,633,478 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 75.06 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -13.47 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20633388 90 + 22407834 AUAGCGACCAAGUUGGCCAUCCUCACUUUCAGUAUUUUUUGCAGAGUUUAUUUGGUGCACAAGCACCGGUAGUCAAUAUAAU-UAUAAUUA ...........((((((...((..((((((((......))).)))))......(((((....)))))))..)))))).....-........ ( -19.20) >DroSec_CAF1 191631 90 + 1 AUAGCGACCAAGUUGGCCAUCCUCACAUUCAGUAUUUUUUGCAGAAUUUAUUUGGUGCACAAGCACCGGUAGUCAAUAUAAU-UAUAAUUA ...........((((((...((....((((.(((.....))).))))......(((((....)))))))..)))))).....-........ ( -18.30) >DroSim_CAF1 184772 90 + 1 AUAGCGACCAAGUUGGCCAUCCUCACAUUCAGUAUUUUUUGCAGAAUUUAUUUGGUGCACAAGCACCGGUAGUCAAUAUAAU-UAUAAUUA ...........((((((...((....((((.(((.....))).))))......(((((....)))))))..)))))).....-........ ( -18.30) >DroYak_CAF1 136904 90 + 1 AAUGCGACCAAGUUGGCCUUCCCUAGUUUCAGUAUUUUUCGCAGAAUUUAUUUGGUGCACUAGCACCGGUUGUCAAUAUAAU-UAUAAUUA ...((((((.....((.....))..............................(((((....))))))))))).........-........ ( -16.10) >DroAna_CAF1 151076 69 + 1 AAACCG--CGAGUUGGCAUCACC---GUUCAC----------------UAUUUGGUGCACAAGCACCGGUUGCCAAUAUAAU-UAUAAUUA ......--...(((((((..(((---(....)----------------.....(((((....))))))))))))))).....-........ ( -19.20) >DroPer_CAF1 214547 72 + 1 AUACCCAACGUGUUGGCAGCUC-------------------CGUAAAUUAUUUGGUGCACAAGCACCGGUUGCCAAUACAAUUUAUAAAUA .........((((((((((((.-------------------............(((((....)))))))))))))))))............ ( -24.81) >consensus AUAGCGACCAAGUUGGCCAUCCUCACAUUCAGUAUUUUUUGCAGAAUUUAUUUGGUGCACAAGCACCGGUAGUCAAUAUAAU_UAUAAUUA ...........((((((.(((..........(((.....)))...........(((((....)))))))).)))))).............. (-13.47 = -14.00 + 0.53)

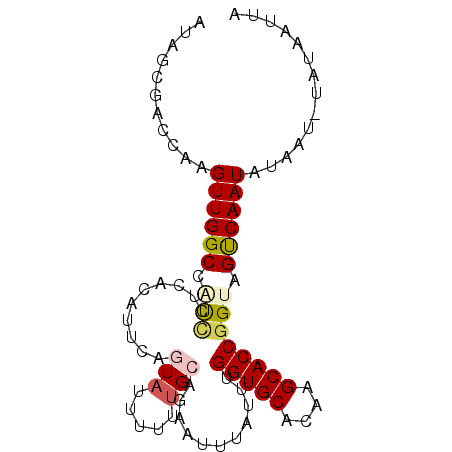
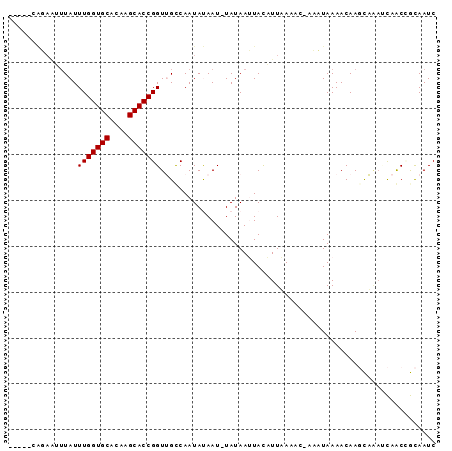
| Location | 20,633,424 – 20,633,515 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -14.82 |
| Consensus MFE | -9.10 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20633424 91 + 22407834 UUUUGCAGAGUUUAUUUGGUGCACAAGCACCGGUAGUCAAUAUAAU-UAUAAUUACAUUAAACC-ACCUAAAACAAGCUGAUCAACCGCAAUC ..((((...(((((..((((((....))))))(((((..(((....-))).)))))..)))))(-(.((......)).)).......)))).. ( -16.00) >DroPse_CAF1 209274 84 + 1 -----CGUAAAUUAUUUGGUGCACAAGCACCGGUUGCCAAUACAAUUUAUAAAUAUUUUUACAC-AAAUAAAACAACUA---UAGCUAUAAUC -----.(((((.((((((((((....)))))(((((......)))))...)))))..)))))..-..............---........... ( -13.70) >DroSim_CAF1 184808 91 + 1 UUUUGCAGAAUUUAUUUGGUGCACAAGCACCGGUAGUCAAUAUAAU-UAUAAUUACAUUAAACC-ACCUAAAACAAGCUGAUCAACCGCAAUC ..((((...........(((((....)))))(((.((.(((.((((-....)))).)))..)).-)))...................)))).. ( -15.30) >DroYak_CAF1 136940 92 + 1 UUUCGCAGAAUUUAUUUGGUGCACUAGCACCGGUUGUCAAUAUAAU-UAUAAUUACAUUAAACCCUCCUAAAACAAGCAAAUCAACGGCAAUC ....((...........(((((....)))))((((...(((.((((-....)))).))).))))............))............... ( -14.10) >DroAna_CAF1 151103 79 + 1 ------------UAUUUGGUGCACAAGCACCGGUUGCCAAUAUAAU-UAUAAUUACUUUUACAC-AAAUAAAACAAGCAAAACAACGGCAAUC ------------.....(((((....)))))(((((((.(((....-)))......(((((...-...))))).............))))))) ( -16.10) >DroPer_CAF1 214569 87 + 1 -----CGUAAAUUAUUUGGUGCACAAGCACCGGUUGCCAAUACAAUUUAUAAAUAUUUUUACAC-AAAUAAAACAACUAAACUAGCUAUAAUC -----.(((((.((((((((((....)))))(((((......)))))...)))))..)))))..-............................ ( -13.70) >consensus _____CAGAAUUUAUUUGGUGCACAAGCACCGGUUGCCAAUAUAAU_UAUAAUUACAUUAAAAC_AAAUAAAACAAGCAAAUCAACCGCAAUC ...............(((((((....)))))))............................................................ ( -9.10 = -9.10 + -0.00)

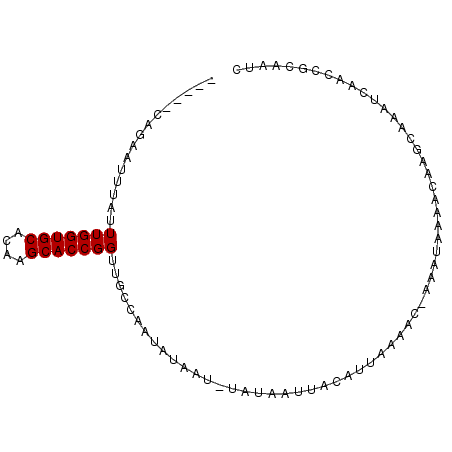
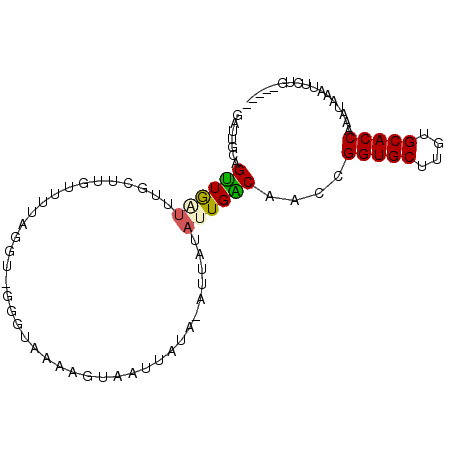
| Location | 20,633,424 – 20,633,515 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.28 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20633424 91 - 22407834 GAUUGCGGUUGAUCAGCUUGUUUUAGGU-GGUUUAAUGUAAUUAUA-AUUAUAUUGACUACCGGUGCUUGUGCACCAAAUAAACUCUGCAAAA ..((((((......((.((((((..(((-((((.((((((((....-)))))))))))))))(((((....))))))))))).)))))))).. ( -30.00) >DroPse_CAF1 209274 84 - 1 GAUUAUAGCUA---UAGUUGUUUUAUUU-GUGUAAAAAUAUUUAUAAAUUGUAUUGGCAACCGGUGCUUGUGCACCAAAUAAUUUACG----- ((((((.....---..((((((.(((..-.((((((....))))))....)))..)))))).(((((....)))))..))))))....----- ( -17.70) >DroSim_CAF1 184808 91 - 1 GAUUGCGGUUGAUCAGCUUGUUUUAGGU-GGUUUAAUGUAAUUAUA-AUUAUAUUGACUACCGGUGCUUGUGCACCAAAUAAAUUCUGCAAAA ..((((((.................(((-((((.((((((((....-)))))))))))))))(((((....))))).........)))))).. ( -29.60) >DroYak_CAF1 136940 92 - 1 GAUUGCCGUUGAUUUGCUUGUUUUAGGAGGGUUUAAUGUAAUUAUA-AUUAUAUUGACAACCGGUGCUAGUGCACCAAAUAAAUUCUGCGAAA ............(((((....((((....(((((((((((((....-))))))))))..)))(((((....)))))...))))....))))). ( -22.60) >DroAna_CAF1 151103 79 - 1 GAUUGCCGUUGUUUUGCUUGUUUUAUUU-GUGUAAAAGUAAUUAUA-AUUAUAUUGGCAACCGGUGCUUGUGCACCAAAUA------------ (.((((((.(((.....((((.((((((-......))))))..)))-)..))).)))))).)(((((....))))).....------------ ( -19.10) >DroPer_CAF1 214569 87 - 1 GAUUAUAGCUAGUUUAGUUGUUUUAUUU-GUGUAAAAAUAUUUAUAAAUUGUAUUGGCAACCGGUGCUUGUGCACCAAAUAAUUUACG----- ((((((.((((((.(((((....(((((-......)))))......))))).))))))....(((((....)))))..))))))....----- ( -20.30) >consensus GAUUGCAGUUGAUUUGCUUGUUUUAGGU_GGGUAAAAGUAAUUAUA_AUUAUAUUGACAACCGGUGCUUGUGCACCAAAUAAAUUCUG_____ .......((((((.......................................))))))....(((((....)))))................. ( -9.56 = -9.28 + -0.28)

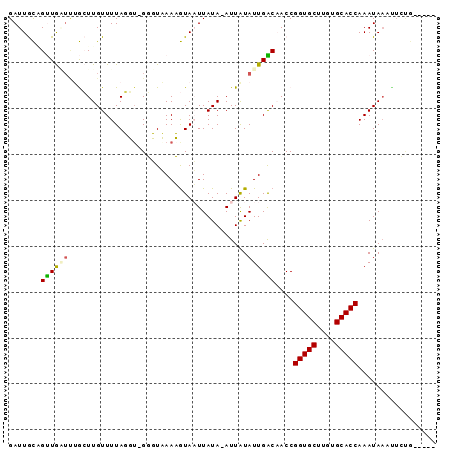
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:49 2006