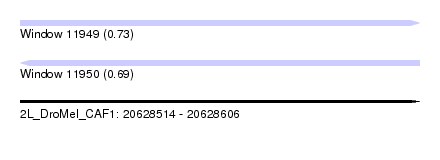
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,628,514 – 20,628,606 |
| Length | 92 |
| Max. P | 0.731646 |

| Location | 20,628,514 – 20,628,606 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 84.60 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -14.98 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20628514 92 + 22407834 GCAUUUCGAACCGCCAAUAUGAAGUUUAUGGCAAUAUGCCUAUUGGCAAAUAUUGGCUACAGUAAGUACAUAGUACAUUUGUACAGAUAUCC ............((((((((...(((...(((.....)))....)))..))))))))........(((((.........)))))........ ( -21.10) >DroSec_CAF1 186812 92 + 1 GCAUUUCGAACUGCCAAUAUGAAGUUUAUGGCAAUAUGCCUGUUGGCAAAUAUUGGCUGCAGUAAGUACAUAGUACAUAUGUACAGAUAUCC (((.........((((((((...(((...(((.....)))....)))..))))))))))).....(((((((.....)))))))........ ( -24.70) >DroSim_CAF1 179216 92 + 1 GCAUUUCGAACUGCCAAUAUGAAGUUUAUGGCAAUAUGCCUGUUGGCAAAUAUUGGCUGCAGUAAGUUCAUUGUACAUAUGUACAGAUAUCC .......(((((((((((((...(((...(((.....)))....)))..)))))))).......))))).((((((....))))))...... ( -24.91) >DroYak_CAF1 131832 77 + 1 GCAUUUAGAAUCACCAAUAUGCAGUUUAUGGCAAUAUGCCUAAUGUCAAAUAUUGGCUACAGUAAGUACAU---------------AUAUUC (.(((((......(((((((.........(((.....))).........)))))))......))))).)..---------------...... ( -12.07) >consensus GCAUUUCGAACCGCCAAUAUGAAGUUUAUGGCAAUAUGCCUAUUGGCAAAUAUUGGCUACAGUAAGUACAUAGUACAUAUGUACAGAUAUCC ............((((((((...(((...(((.....)))....)))..))))))))...............((((....))))........ (-14.98 = -15.73 + 0.75)

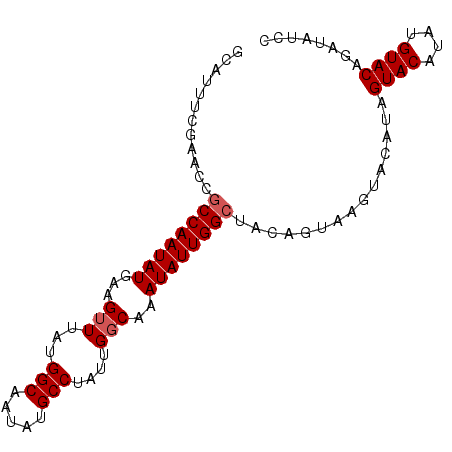
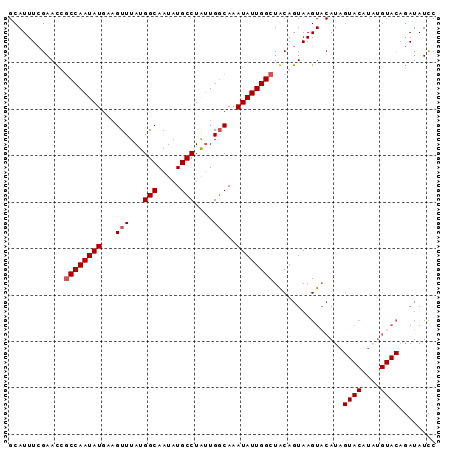
| Location | 20,628,514 – 20,628,606 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 84.60 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -16.61 |
| Energy contribution | -16.30 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20628514 92 - 22407834 GGAUAUCUGUACAAAUGUACUAUGUACUUACUGUAGCCAAUAUUUGCCAAUAGGCAUAUUGCCAUAAACUUCAUAUUGGCGGUUCGAAAUGC ((((.....((((...((((...))))....))))((((((((.........(((.....))).........)))))))).))))....... ( -19.47) >DroSec_CAF1 186812 92 - 1 GGAUAUCUGUACAUAUGUACUAUGUACUUACUGCAGCCAAUAUUUGCCAACAGGCAUAUUGCCAUAAACUUCAUAUUGGCAGUUCGAAAUGC ((((....(((((((.....)))))))........((((((((.........(((.....))).........)))))))).))))....... ( -22.07) >DroSim_CAF1 179216 92 - 1 GGAUAUCUGUACAUAUGUACAAUGAACUUACUGCAGCCAAUAUUUGCCAACAGGCAUAUUGCCAUAAACUUCAUAUUGGCAGUUCGAAAUGC .......(((((....))))).((((((.......((((((((.........(((.....))).........))))))))))))))...... ( -21.18) >DroYak_CAF1 131832 77 - 1 GAAUAU---------------AUGUACUUACUGUAGCCAAUAUUUGACAUUAGGCAUAUUGCCAUAAACUGCAUAUUGGUGAUUCUAAAUGC ((((..---------------...(((.....)))((((((((.........(((.....))).........)))))))).))))....... ( -13.87) >consensus GGAUAUCUGUACAUAUGUACUAUGUACUUACUGCAGCCAAUAUUUGCCAACAGGCAUAUUGCCAUAAACUUCAUAUUGGCAGUUCGAAAUGC ((((....((((....))))...............((((((((.........(((.....))).........)))))))).))))....... (-16.61 = -16.30 + -0.31)

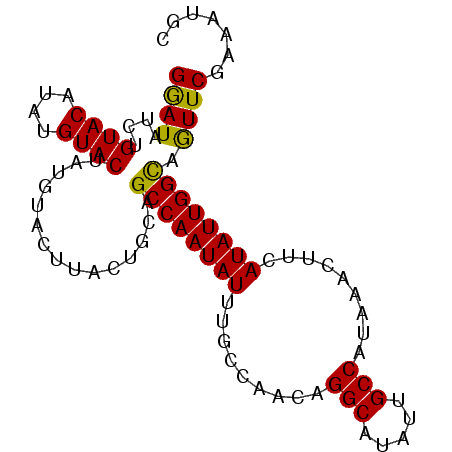
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:45 2006