
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,619,495 – 20,619,588 |
| Length | 93 |
| Max. P | 0.839139 |

| Location | 20,619,495 – 20,619,588 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.34 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20619495 93 - 22407834 CUUGGGA---GGAUUGUGG---------------AGAU--UGGAAA-------GCGGAGGGAUUUGGAAUGCAAGAUGCCAGACGGUGUAAAAUUUGCAUUUUUCUCCGGACAGGGCAAC (((((((---(((..(((.---------------((((--(....(-------.((..((.((((........)))).))...)).)....))))).)))..))))))...))))..... ( -24.90) >DroSec_CAF1 138367 73 - 1 CUUGGGA---G------------------------GAU--UGGAAA-------GCGGAAGGAGAUGGAAUGCAAGAUGCCAGACGGUGUAAAAUUUGCAUUUUUCUCCG----------- .......---.------------------------...--......-------......(((((.(((((((((((((((....)))))....))))))))))))))).----------- ( -20.00) >DroSim_CAF1 135835 82 - 1 CUUGGGA---GGAUUGGGA---------------GGAU--UGGAAA-------GCGGAAGGAGAUGGAAUGCAAGAUGCCAGACGGUGUAAAAUUUGCAUUUUUCUCCG----------- ((..(..---...)..)).---------------....--......-------......(((((.(((((((((((((((....)))))....))))))))))))))).----------- ( -21.40) >DroEre_CAF1 123189 111 - 1 CUUGGGACCGGGAUUGGGACCGGGAUUGGGAUUGGGAU--UGCGGA-------GAGGAAGGAGAUGGAAUGCAAGAUGCCAGACGGUGUAAAAUUAGCAUUUUUCUCGAGAGAAAGCCAC ((..(..((((........))))..)..))..(((..(--(.(...-------..).)).((((.(((((((((.(((((....)))))....)).))))))))))).........))). ( -28.80) >DroYak_CAF1 128016 83 - 1 CUUGGA----------------------------GGAU--UGAAAA-------GAAGAUGGAGAUGGAAUGCAAGAUGCCAGACGGUGUAAAAUUUGCAUUUUUCUCUGAAGAGAGCAAC ......----------------------------....--......-------......(((((.(((((((((((((((....)))))....)))))))))))))))............ ( -17.30) >DroAna_CAF1 133289 97 - 1 ----AGA---GGAUGGUGA---------------AGCCCCAGGGAAGAGUCAGGGGUCAGGGGGAGAAAUGCAAGAUGCCAGGCGGUGCUAAAUUUACAUUUUUCUUCUUACCCUG-AAC ----...---((..((...---------------..)))).........((((((...(((((((((((((..(((((((....))).....)))).))))))))))))).)))))-).. ( -26.60) >consensus CUUGGGA___GGAUUG_GA_______________GGAU__UGGAAA_______GCGGAAGGAGAUGGAAUGCAAGAUGCCAGACGGUGUAAAAUUUGCAUUUUUCUCCG_A_A__G_AAC ...........................................................(((((.(((((((((((((((....)))))....)))))))))))))))............ (-14.90 = -15.27 + 0.37)

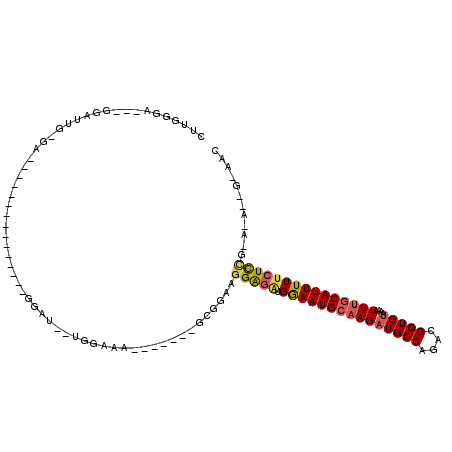
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:39 2006