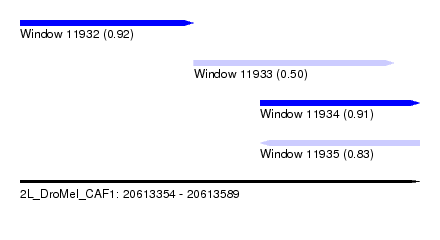
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,613,354 – 20,613,589 |
| Length | 235 |
| Max. P | 0.916475 |

| Location | 20,613,354 – 20,613,456 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 64.99 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -4.90 |
| Energy contribution | -6.23 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.19 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20613354 102 + 22407834 AGUAGGCCAAAACAAAAAGCCUGCAAGGCAACAAAGACACGCACACAU----UGGUGCCUGUGAGGCACACUCAUGUGCAUUGUCCAUGCACAGAUGCGCCAGCCC .((((((...........))))))..(((.......(((.((((....----..)))).)))..(((.((.((.(((((((.....))))))))))).))).))). ( -37.70) >DroPse_CAF1 190028 83 + 1 CAGAAAAUAUACACAAAAGCCUGCAGGGCAACAAAGGG-UGCACACAU----C-------------CACAUCCACAUCCACUG-----GCACAUUUGCAUCCACAC ..................(((.....)))......(((-((((.....----(-------------((.............))-----)......))))))).... ( -15.52) >DroSec_CAF1 132397 102 + 1 AGUAGGCCAAAACAAAAAGCCUGCCAGGCAACAAAGCCACGCACACAU----UGGUGCCUGUGACGCACACUCAUGUGCAUUGUCCAUGCACAGAUGCGCCCACCC .((((((...........))))))..(((......)))..((((....----..))))..(((.((((...((.(((((((.....)))))))))))))..))).. ( -32.50) >DroSim_CAF1 128407 102 + 1 AGUAGGCCAAAACAAAAAGCCUGCCAGGCAACAAAGGCACGCACACAU----CGGUGCCUGUGACGCACACUCAUGUGCAUUGUCCAUGCACAGAUGCGCCCACCC .((.(((...........((.((((.(....)...)))).))...(((----(.((((.((.((((((((....)))))...))))).)))).)))).))).)).. ( -34.90) >DroYak_CAF1 122057 89 + 1 AGAAUUCCAAAACAAAAAGCCUACCAGGCAACAAAGGCUCGCCCACACAGAGUAGUGUCUUCAACGCACACACAU--GCAUUGUCCACACA--------------- ..................(((.....))).......((((.........)))).((((...(((.(((......)--)).)))...)))).--------------- ( -16.60) >DroPer_CAF1 195706 83 + 1 CAGAAAAUAUACACAAAAGCCUGCAGGGCAACAAAGGG-UGCACACAU----C-------------CACAUCCACAUCCACUG-----GCACAUUUGCAUCCACAC ..................(((.....)))......(((-((((.....----(-------------((.............))-----)......))))))).... ( -15.52) >consensus AGUAGGCCAAAACAAAAAGCCUGCAAGGCAACAAAGGCACGCACACAU____CGGUGCCUGUGACGCACACUCAUGUGCAUUGUCCAUGCACAGAUGCGCCCACCC ..................(((.....)))..........((((...........((((.......))))............((......))....))))....... ( -4.90 = -6.23 + 1.33)

| Location | 20,613,456 – 20,613,574 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.85 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
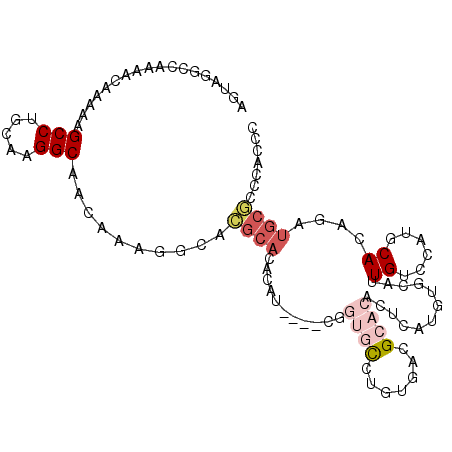
>2L_DroMel_CAF1 20613456 118 + 22407834 A-CUCACACUCACACACACACGUGCAUACCACUCGGAGGAAUUGUCAUUAUACAAUGCAACCGUGAAUAAUAGGCUGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAGCC .-.((((.....(((......)))..........((..(.(((((......))))).)..))))))......(((((.....((((.(((.((((.....)))).))).)))).))))) ( -29.30) >DroSec_CAF1 132499 112 + 1 U-CUCUCACACACACACA---ACACACACUACUCGGAGAAAUUGUCAUCAUACAAUGCAACCGUGAAUAAAAGGCUGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCA--- .-................---..........(.(((....(((((......)))))....))).).......((.....)).((((.(((.((((.....)))).))).))))...--- ( -17.90) >DroSim_CAF1 128509 115 + 1 U-CUCUCACACACACACA---ACACACACCACUCGGAGGAAUUGUCAUUAUACAAUGCAACCGUGAAUAAUAGGCUGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAGCC .-...((((.........---.......((....)).((..(((.((((....)))))))))))))......(((((.....((((.(((.((((.....)))).))).)))).))))) ( -27.00) >DroEre_CAF1 118143 99 + 1 ----------------C----GCGCACACCACUCGGAGGAAUUGUCAUUAAACAAUGCACCCGUGAAUAAUGGGCUGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAGCC ----------------.----((((......(.(((..(.(((((......))))).)..))).).....((((....))))((((.(((.((((.....)))).))).)))))).)). ( -27.90) >DroYak_CAF1 122146 113 + 1 --CUCUCACACGCUCGU----GAACACACCACUCGGAGGAAUUGUCAUUAUACAAUGCAACCGUGAAUAAUAGGCUGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAGCC --...((((......((----(.......)))..((..(.(((((......))))).)..))))))......(((((.....((((.(((.((((.....)))).))).)))).))))) ( -29.40) >DroAna_CAF1 126977 113 + 1 UACCCACCUACACACAUA---CCACA---CACUCGCACGAAUUGUCAUUAUACAAUGCAUCCGUAAAUAAUAGGCUGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAAGC ..................---.....---.....(((((...((.((((....))))))..)))..................((((.(((.((((.....)))).))).)))))).... ( -17.10) >consensus U_CUCUCACACACACACA___GCACACACCACUCGGAGGAAUUGUCAUUAUACAAUGCAACCGUGAAUAAUAGGCUGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAGCC ...............................(.(((....(((((......)))))....))).).......(((((.....((((.(((.((((.....)))).))).)))).))))) (-19.68 = -20.85 + 1.17)


| Location | 20,613,495 – 20,613,589 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -13.95 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
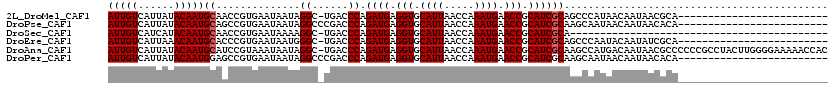
>2L_DroMel_CAF1 20613495 94 + 22407834 AUUGUCAUUAUACAAUGCAACCGUGAAUAAUAGGC-UGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAGCCCAUAACAAUAACGCA------------------------- .(((.((((....))))))).(((........(((-((.....((((.(((.((((.....)))).))).)))).)))))..........)))..------------------------- ( -21.47) >DroPse_CAF1 190125 95 + 1 AUUGUCAUUAUACAAUGCAGCCGUGAAUAAUAGGCCCGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAAGCAAUAACAAUAACACA------------------------- (((((.(((......(((.(((..........)))........((((.(((.((((.....)))).))).)))))))...))).)))))......------------------------- ( -18.80) >DroSec_CAF1 132535 76 + 1 AUUGUCAUCAUACAAUGCAACCGUGAAUAAAAGGC-UGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCA------------------------------------------- (((((......)))))((..............((.-....)).((((.(((.((((.....)))).))).)))))).------------------------------------------- ( -14.90) >DroEre_CAF1 118163 94 + 1 AUUGUCAUUAAACAAUGCACCCGUGAAUAAUGGGC-UGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAGCCCAAUACAAUAUCGCA------------------------- (((((......)))))......((((.((.(((((-((.....((((.(((.((((.....)))).))).)))).))))))).)).....)))).------------------------- ( -27.40) >DroAna_CAF1 127011 119 + 1 AUUGUCAUUAUACAAUGCAUCCGUAAAUAAUAGGC-UGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAAGCCAUGACAAUAACGCCCCCCGCCUACUUGGGGAAAAACCAC ((((((((.......(((....))).......(((-(......((((.(((.((((.....)))).))).))))...))))))))))))......(((((......)))))......... ( -32.30) >DroPer_CAF1 195803 95 + 1 AUUGUCAUUAUACAAUGGAGCCGUGAAUAAUAGGCCCGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAAGCAAUAACAAUAACACA------------------------- (((((.(((......(((.(((..........)))))).....((((.(((.((((.....)))).))).))))......))).)))))......------------------------- ( -20.90) >consensus AUUGUCAUUAUACAAUGCAACCGUGAAUAAUAGGC_UGACCCAGAUGAGGUGCAUUAACCAAAUGAACCGCAUCGCAAGCCAUAACAAUAACGCA_________________________ (((((......)))))((..............((......)).((((.(((.((((.....)))).))).))))))............................................ (-13.95 = -14.12 + 0.17)
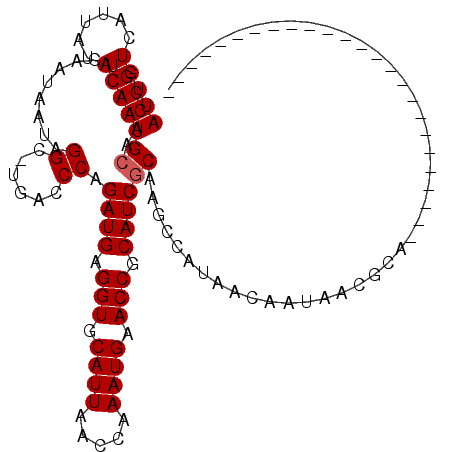
| Location | 20,613,495 – 20,613,589 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -19.45 |
| Energy contribution | -21.40 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
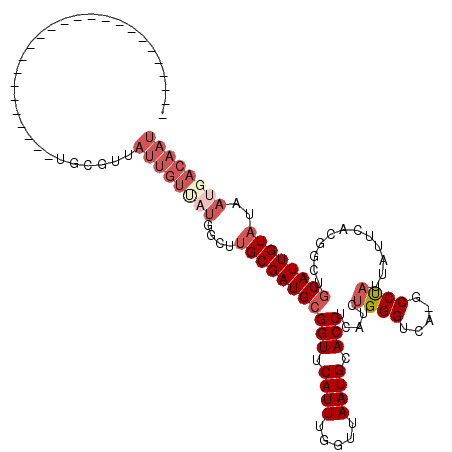
>2L_DroMel_CAF1 20613495 94 - 22407834 -------------------------UGCGUUAUUGUUAUGGGCUGCGAUGCGGUUCAUUUGGUUAAUGCACCUCAUCUGGGUCA-GCCUAUUAUUCACGGUUGCAUUGUAUAAUGACAAU -------------------------...((((((((.(((((((((((((.(((.((((.....)))).))).))))...).))-)))))).....(((((...)))))))))))))... ( -26.50) >DroPse_CAF1 190125 95 - 1 -------------------------UGUGUUAUUGUUAUUGCUUGCGAUGCGGUUCAUUUGGUUAAUGCACCUCAUCUGGGUCGGGCCUAUUAUUCACGGCUGCAUUGUAUAAUGACAAU -------------------------......((((((((((..((((((((((((....((..(((((..(((.((....)).)))..)))))..)).)))))))))))))))))))))) ( -28.00) >DroSec_CAF1 132535 76 - 1 -------------------------------------------UGCGAUGCGGUUCAUUUGGUUAAUGCACCUCAUCUGGGUCA-GCCUUUUAUUCACGGUUGCAUUGUAUGAUGACAAU -------------------------------------------((((((((((..((...(((......))).....))..))(-(((..........)))))))))))).......... ( -17.40) >DroEre_CAF1 118163 94 - 1 -------------------------UGCGAUAUUGUAUUGGGCUGCGAUGCGGUUCAUUUGGUUAAUGCACCUCAUCUGGGUCA-GCCCAUUAUUCACGGGUGCAUUGUUUAAUGACAAU -------------------------((((.....(((.((((((((((((.(((.((((.....)))).))).))))...).))-))))).))).......))))(((((....))))). ( -29.30) >DroAna_CAF1 127011 119 - 1 GUGGUUUUUCCCCAAGUAGGCGGGGGGCGUUAUUGUCAUGGCUUGCGAUGCGGUUCAUUUGGUUAAUGCACCUCAUCUGGGUCA-GCCUAUUAUUUACGGAUGCAUUGUAUAAUGACAAU .......((((((........))))))....((((((((....(((((((((((.((((.....)))).))).....((((...-.))))............))))))))..)))))))) ( -35.30) >DroPer_CAF1 195803 95 - 1 -------------------------UGUGUUAUUGUUAUUGCUUGCGAUGCGGUUCAUUUGGUUAAUGCACCUCAUCUGGGUCGGGCCUAUUAUUCACGGCUCCAUUGUAUAAUGACAAU -------------------------......((((((((((..(((((((.(((.((((.....)))).))).))))(((((((.............)))).)))..))))))))))))) ( -26.02) >consensus _________________________UGCGUUAUUGUUAUGGCUUGCGAUGCGGUUCAUUUGGUUAAUGCACCUCAUCUGGGUCA_GCCUAUUAUUCACGGCUGCAUUGUAUAAUGACAAU ...............................((((((((....(((((((((((.((((.....)))).))).....((((.....))))............))))))))..)))))))) (-19.45 = -21.40 + 1.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:32 2006