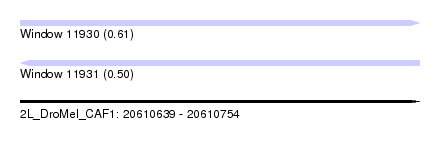
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,610,639 – 20,610,754 |
| Length | 115 |
| Max. P | 0.611860 |

| Location | 20,610,639 – 20,610,754 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -21.45 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
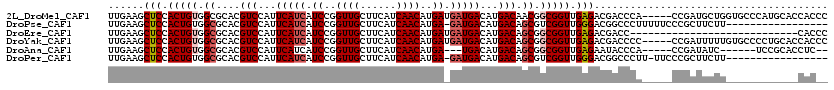
>2L_DroMel_CAF1 20610639 115 + 22407834 UUGAAGCUCCACUGUGGCGCACGUCCAUUCAUCAUCCGGUUGCUUCAUCAACAUGAUGAUGACAUGACAACGGCGGUUGAGACGACCCA-----CCGAUGCUGGUGCCCAUGCACCACCC .....((.((.....)).))..(((...((((((((..((((......))))..))))))))...)))..(((.(((((...)))))..-----)))....((((((....))))))... ( -37.30) >DroPse_CAF1 186327 102 + 1 UUGAAGCUCCACUGUGGCGCACGUCCAUUCAUCAUCCGGUUGCUUCAUCAACAUGA-GAUGACAUGACAGCGUCGGUUGGGACGGCCCUUUUUCCCGCUUCUU----------------- ..(((((........((.((.(((((..((((((((((((((......)))).)).-)))))..)))((((....)))))))))))))........)))))..----------------- ( -32.19) >DroEre_CAF1 115669 92 + 1 UUGAAGCUCCACUGUGGCGCACGUCCAUUCAUCAUCCGGUUGCUUCAUCAACAUGAUGAUGACAUGACAGCGGCGGUUGAGACGACC----------------------------CACCC .....((((.(((((.((....(((...((((((((..((((......))))..))))))))...))).)).))))).))).)....----------------------------..... ( -29.80) >DroYak_CAF1 119395 115 + 1 UUGAAGCUCCACUGUGGCGCACGUCCAUUCAUCAUCCGGUUGCUUCAUCAACAUGAUGAUGACAUGACAGCGGCGGUUGAGACGACCCC-----CCGAUUUUUGUGCCCCUGCACCACCC .............((((.(((.(((...((((((((..((((......))))..))))))))...)))...((.(((.((((((.....-----.)).))))...)))))))).)))).. ( -33.80) >DroAna_CAF1 124405 104 + 1 UUGAAGCUCCACUGUGGCGCACGUCCAUUCAUCAUCCGGUUGCUUCAUCAACAUGA---UGACAUGACAGCGGCGGUUGAGAAUACCCA-----CCGAUAUC------UCCGCACCUC-- .....((.((.....)).))..(((...(((((((...((((......))))))))---)))...))).((((((((.(.......).)-----))).....------.)))).....-- ( -25.50) >DroPer_CAF1 192022 101 + 1 UUGAAGCUCCACUGUGGCGCACGUCCAUUCAUCAUCCGGUUGCUUCAUCAACAUGA-GAUGACAUGACAGCGUCGGUUGGGACGGCCCUU-UUCCCGCUUCUU----------------- ..(((((........((.((.(((((..((((((((((((((......)))).)).-)))))..)))((((....)))))))))))))..-.....)))))..----------------- ( -31.76) >consensus UUGAAGCUCCACUGUGGCGCACGUCCAUUCAUCAUCCGGUUGCUUCAUCAACAUGA_GAUGACAUGACAGCGGCGGUUGAGACGACCCA_____CCGAUUCUU____________CAC__ ......(((.(((((.((....(((...(((((.((..((((......))))..)).)))))...))).)).))))).)))....................................... (-21.45 = -22.07 + 0.61)
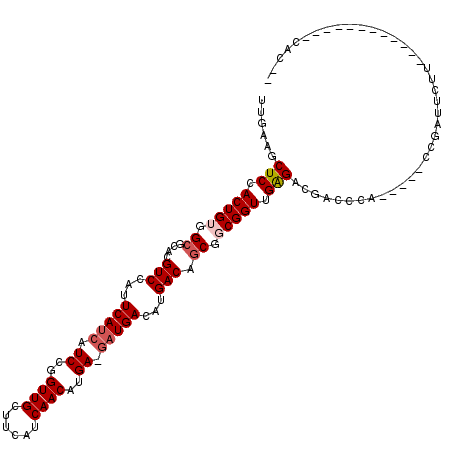

| Location | 20,610,639 – 20,610,754 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
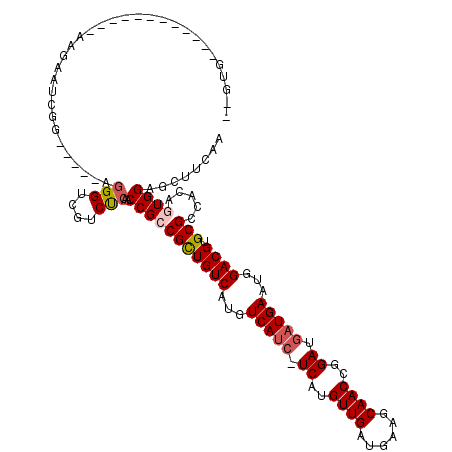
>2L_DroMel_CAF1 20610639 115 - 22407834 GGGUGGUGCAUGGGCACCAGCAUCGG-----UGGGUCGUCUCAACCGCCGUUGUCAUGUCAUCAUCAUGUUGAUGAAGCAACCGGAUGAUGAAUGGACGUGCGCCACAGUGGAGCUUCAA ..((((((((((....((((((.(((-----(((..........)))))).)))....((((((((..((((......))))..)))))))).))).))))))))))............. ( -46.90) >DroPse_CAF1 186327 102 - 1 -----------------AAGAAGCGGGAAAAAGGGCCGUCCCAACCGACGCUGUCAUGUCAUC-UCAUGUUGAUGAAGCAACCGGAUGAUGAAUGGACGUGCGCCACAGUGGAGCUUCAA -----------------..(((((((((..........))))..(((.(((((((...(((((-((..((((......))))..)).)))))...)))).)))......))).))))).. ( -32.90) >DroEre_CAF1 115669 92 - 1 GGGUG----------------------------GGUCGUCUCAACCGCCGCUGUCAUGUCAUCAUCAUGUUGAUGAAGCAACCGGAUGAUGAAUGGACGUGCGCCACAGUGGAGCUUCAA ((.((----------------------------((....)))).)).(((((((((((((.(((((((((((......))))...)))))))...))))))....)))))))........ ( -32.10) >DroYak_CAF1 119395 115 - 1 GGGUGGUGCAGGGGCACAAAAAUCGG-----GGGGUCGUCUCAACCGCCGCUGUCAUGUCAUCAUCAUGUUGAUGAAGCAACCGGAUGAUGAAUGGACGUGCGCCACAGUGGAGCUUCAA ..(((((((((.(((.(.......)(-----(..(......)..))))).))(((...((((((((..((((......))))..))))))))...)))..)))))))............. ( -41.40) >DroAna_CAF1 124405 104 - 1 --GAGGUGCGGA------GAUAUCGG-----UGGGUAUUCUCAACCGCCGCUGUCAUGUCA---UCAUGUUGAUGAAGCAACCGGAUGAUGAAUGGACGUGCGCCACAGUGGAGCUUCAA --(((((...((------((((((..-----..)))).))))..(((((((((((...(((---((((((((......))))...)))))))...)))).))).....)))).))))).. ( -34.30) >DroPer_CAF1 192022 101 - 1 -----------------AAGAAGCGGGAA-AAGGGCCGUCCCAACCGACGCUGUCAUGUCAUC-UCAUGUUGAUGAAGCAACCGGAUGAUGAAUGGACGUGCGCCACAGUGGAGCUUCAA -----------------..(((((((((.-........))))..(((.(((((((...(((((-((..((((......))))..)).)))))...)))).)))......))).))))).. ( -33.00) >consensus __GUG____________AAGAAUCGG_____AGGGUCGUCUCAACCGCCGCUGUCAUGUCAUC_UCAUGUUGAUGAAGCAACCGGAUGAUGAAUGGACGUGCGCCACAGUGGAGCUUCAA ................................(((....)))..(((((((((((...(((((.((..((((......))))..)).)))))...)))).))).....))))........ (-22.24 = -22.72 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:29 2006