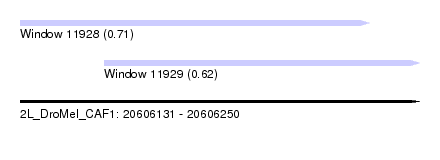
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,606,131 – 20,606,250 |
| Length | 119 |
| Max. P | 0.706189 |

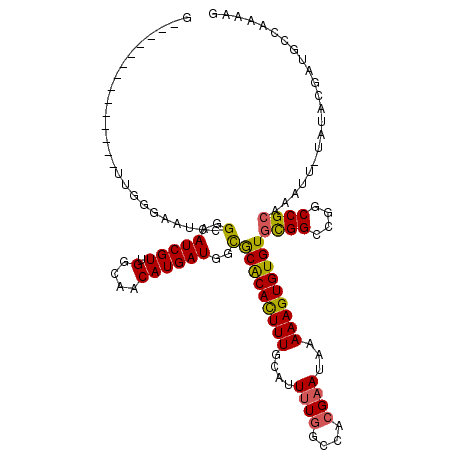
| Location | 20,606,131 – 20,606,235 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -23.09 |
| Energy contribution | -22.90 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20606131 104 + 22407834 G--------------UUGGGAAUCCGGAAUCGUUGGCAACAUGAUGGUGCACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGUGGCCGGCCGCAAAUU-UAUACGAUGCCAAAAG .--------------..((....))((.(((((((((........((((((.....)))))).((((((((.(((.....))).))))))))))))......-...))))).))..... ( -29.20) >DroSec_CAF1 125283 104 + 1 G--------------UUGCGAAUCCGGAAUCGUUGGCAACAUGAUGGCACACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGCAAAUU-UAUACGAUGCCAAAAG .--------------(((((...((((.(((((.(....)))))).(((((((((((....((((....))))...)))))))))))..)))).)))))...-................ ( -36.70) >DroSim_CAF1 121267 104 + 1 G--------------UUGCGAAUCCGGAAUCGUUGGCAACAUGAUGGCGCACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGGAAAUU-UAUACGAUGCCAAAAG .--------------.......(((((.(((((.(....)))))).(((((((((((....((((....))))...)))))))))))......)))))....-................ ( -32.70) >DroEre_CAF1 111444 117 + 1 GAGCGGAGAUCGGAAUCCGGAAUCCGGAAUCGUUGGCAACAUGAUGGCGCACACUUUGCAUU-UGGCCACGAAUAAAAAGUGUGUGCGGCAGGCCGCAAAUU-UAUACGCUGGCCAGAG ..(((((..((((...))))..)))...(((((.(....)))))).(((((((((((..(((-((....)))))..))))))))))).)).((((((.....-.....)).)))).... ( -44.30) >DroYak_CAF1 114953 104 + 1 G--------------UUCGGAAAUCGAAAUCGUUGGCAGCAUGAUGGCGCGCACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCAGGCCGCAAAUU-UAUAGAAUGGCCAGAG .--------------((((.....))))....(((((.((......))(((((((((....((((....))))...)))))))))((((....)))).....-.........))))).. ( -30.50) >DroAna_CAF1 118297 101 + 1 G--------------GUG----GCCGAAAUCGUUGGCAACAUGAUGGCGCACAUUUUACAUUUUGGCCAAGAAUGAAAAGUGUGUGCGGCCCGCCGCAAAUAACAUUUAAUGGGAUAAA (--------------(((----(((...(((((.(....)))))).(((((((((((.((((((.....)))))).))))))))))))).)))))........................ ( -36.80) >consensus G______________UUGGGAAUCCGGAAUCGUUGGCAACAUGAUGGCGCACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGCAAAUU_UAUACGAUGCCAAAAG .........................(..(((((.(....))))))..)(((((((((....((((....))))...)))))))))((((....))))...................... (-23.09 = -22.90 + -0.19)

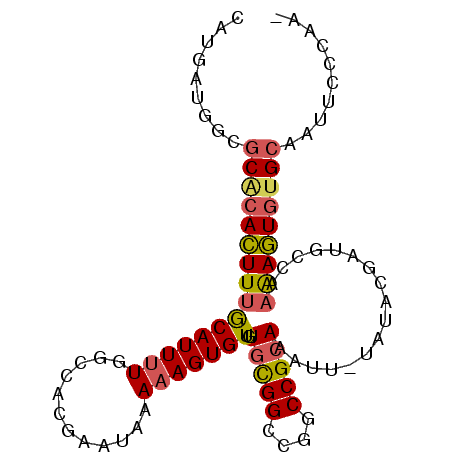
| Location | 20,606,156 – 20,606,250 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.38 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20606156 94 + 22407834 CAUGAUGGUGCACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGUGGCCGGCCGCAAAUU-UAUACGAUGCCAAAAGUGUGCAAUUCCCAAA .....(((((((((((((.......(((((((.(((.....))).)))))))(((((......-....))..))).))))))))))....))).. ( -31.50) >DroSec_CAF1 125308 94 + 1 CAUGAUGGCACACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGCAAAUU-UAUACGAUGCCAAAAGUGUGCAAUUCCCCAA (((..(((((((((((((....((((....))))...))))))))(((((....)))))....-.......)))))...)))............. ( -28.30) >DroSim_CAF1 121292 94 + 1 CAUGAUGGCGCACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGGAAAUU-UAUACGAUGCCAAAAGUGUGCAAUUCCCCCC .......(((((((((((....((((....))))...)))))))))))((....))((((.(.-(((((..........))))).).)))).... ( -27.30) >DroEre_CAF1 111483 92 + 1 CAUGAUGGCGCACACUUUGCAUU-UGGCCACGAAUAAAAAGUGUGUGCGGCAGGCCGCAAAUU-UAUACGCUGGCCAGAGUGUGCAAUACUCAA- ..((.(.(((((((((((..(((-((....)))))..))))))))))).)))((((((.....-.....)).)))).((((((...))))))..- ( -35.70) >DroYak_CAF1 114978 93 + 1 CAUGAUGGCGCGCACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCAGGCCGCAAAUU-UAUAGAAUGGCCAGAGUGUGCAAUUCUUAA- ...((..((((((.....))((((((((((........((((...(((((....))))).)))-)......))))))))))))))...))....- ( -30.74) >DroAna_CAF1 118318 95 + 1 CAUGAUGGCGCACAUUUUACAUUUUGGCCAAGAAUGAAAAGUGUGUGCGGCCCGCCGCAAAUAACAUUUAAUGGGAUAAAUUCGCAGUACUUAUU ...(((((((((((((((.((((((.....)))))).)))))))))))((....))........))))...((.((.....)).))......... ( -25.80) >consensus CAUGAUGGCGCACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGCAAAUU_UAUACGAUGCCAAAAGUGUGCAAUUCCCAA_ .........((((((((((((((((............))))))).(((((....))))).................))))))))).......... (-21.27 = -21.38 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:27 2006