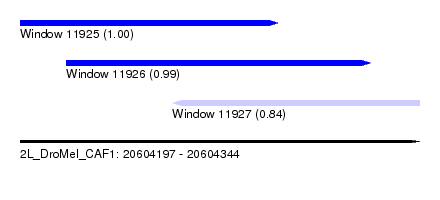
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,604,197 – 20,604,344 |
| Length | 147 |
| Max. P | 0.999238 |

| Location | 20,604,197 – 20,604,292 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.23 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
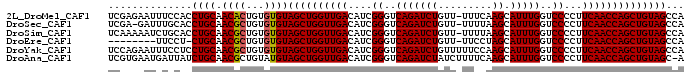
>2L_DroMel_CAF1 20604197 95 + 22407834 UCGAGAAUUUCCACCUGCAACACUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUU-UUUCAAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCA ..............((((.((((...))))((((((((((....((..(((((((...-.....)).)))))..))...))))))))))))))... ( -31.30) >DroSec_CAF1 123428 94 + 1 UCGA-GAUUUGCACCUGCAACGCUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUU-UUUUAAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCA ....-...((((....)))).((((....(((((((((((....((..(((((((...-.....)).)))))..))...))))))))))))))).. ( -33.00) >DroSim_CAF1 119381 95 + 1 UCAAAAAUCUGCACCUGCAACGCUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUU-UUUUAAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCA .........(((....)))..((((....(((((((((((....((..(((((((...-.....)).)))))..))...))))))))))))))).. ( -32.30) >DroEre_CAF1 109752 86 + 1 --------UUCCU-CUGCAACGCUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUU-UUCCUAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCA --------.....-((((.((((...))))((((((((((....((..((((((((..-....))).)))))..))...))))))))))))))... ( -30.80) >DroYak_CAF1 113081 96 + 1 UCCAGAAUUUCCUCCUGCAACGCUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUUUUUCCAAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCA ..............((((.((((...))))((((((((((....((..(((((((.........)).)))))..))...))))))))))))))... ( -30.10) >DroAna_CAF1 116864 95 + 1 UCGUGAAUGAUUAUCUGCAACGCUGUAUGUAGCUGGUUGACAUCGGGUCAGAUCUAUCUUUUCAAGCAUUUGGUCCCCUUCAACCAGCUGUAGC-A .....................((((....(((((((((((....((..(((((((.........)).)))))..))...)))))))))))))))-. ( -29.40) >consensus UCGAGAAUUUCCACCUGCAACGCUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUU_UUUCAAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCA ..............((((.((((...))))((((((((((....((..(((((((.........)).)))))..))...))))))))))))))... (-29.20 = -29.23 + 0.03)
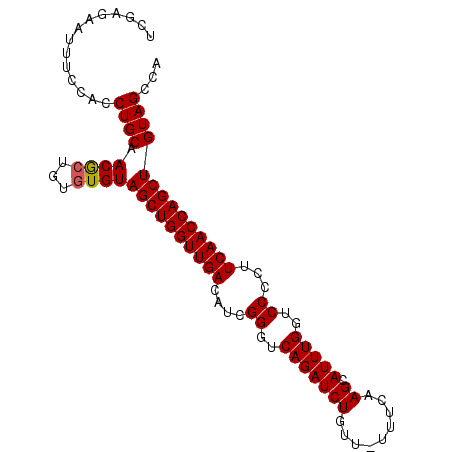
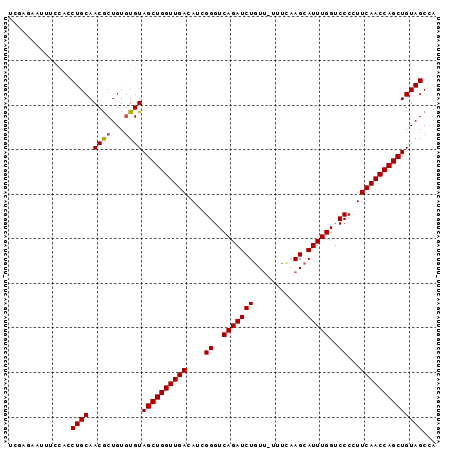
| Location | 20,604,214 – 20,604,326 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.71 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -35.52 |
| Energy contribution | -35.55 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20604214 112 + 22407834 CAAC-ACUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUU-UUUCAAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCAGUGGUUGAUGACAACGAAAGCCUAGUGA------ACUGCG ...(-((((..(((((((((((((....((..(((((((...-.....)).)))))..))...)))))))))))))..)))))((((....))))....((..((...------.)))). ( -40.70) >DroPse_CAF1 173378 119 + 1 CAACGGCUGUAUGUAGCUGGUUGACAUCGGGUCAGAUCUAUUUUAUCAAGUAUUUGGUCCCCUUCAACCAGCUGUAU-CAGUGGUUGAUUCCAACAAAAAACAAAAGGUGCCAACAAAUA ((((.((((.((((((((((((((....((..(((((((.........)).)))))..))...))))))))))))))-)))).))))................................. ( -38.90) >DroEre_CAF1 109760 112 + 1 CAAC-GCUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUU-UUCCUAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCAGUGGUUGAUGCCAACUAAAUUCGAGUGA------ACUGUG ...(-((((..(((((((((((((....((..((((((((..-....))).)))))..))...)))))))))))))..)))))((((....))))........((...------.))... ( -38.90) >DroYak_CAF1 113098 113 + 1 CAAC-GCUGUGUGUAGCUGGUUGACAUCGGGUCAGAUCUGUUUUUCCAAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCAGUGGUUGAUGACAACGAAGUACGAGUAA------ACUGUG ...(-((((..(((((((((((((....((..(((((((.........)).)))))..))...)))))))))))))..)))))((((....)))).............------...... ( -37.60) >DroAna_CAF1 116881 112 + 1 CAAC-GCUGUAUGUAGCUGGUUGACAUCGGGUCAGAUCUAUCUUUUCAAGCAUUUGGUCCCCUUCAACCAGCUGUAGC-AGUGGUUGAUGACAACAAAGUACAAGUGA------AAAGUG ...(-(((((.(((((((((((((....((..(((((((.........)).)))))..))...)))))))))))))))-))))((((....)))).............------...... ( -36.80) >DroPer_CAF1 179293 119 + 1 CAACGGCUGUAUGUAGCUGGUUGACAUCGGGUCAGAUCUAUUUUAUCAAGUAUUUGGUCCCCUUCAACCAGCUGUAU-CAGUGGUUGAUUCCAACAAAAAACAAAAGGCGCCAACAAAUA ((((.((((.((((((((((((((....((..(((((((.........)).)))))..))...))))))))))))))-)))).))))................................. ( -38.90) >consensus CAAC_GCUGUAUGUAGCUGGUUGACAUCGGGUCAGAUCUAUUUUUUCAAGCAUUUGGUCCCCUUCAACCAGCUGUAGCCAGUGGUUGAUGACAACAAAAUACAAGUGA______AAAGUG ((((.((((..(((((((((((((....((..(((((((.........)).)))))..))...)))))))))))))..)))).))))................................. (-35.52 = -35.55 + 0.03)

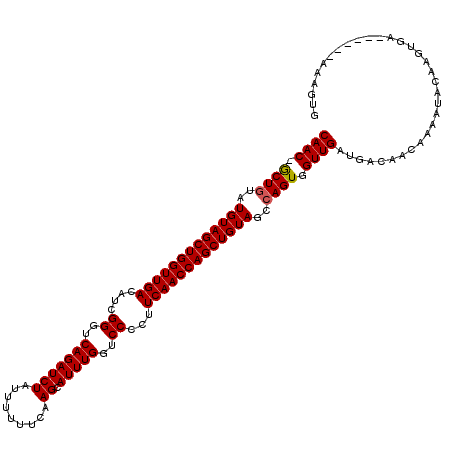
| Location | 20,604,253 – 20,604,344 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.95 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20604253 91 - 22407834 UG----GAAAAUGGUAAU------------------CCAUCGCAGUUCACUAGGCUUUCGUUGUCAUCAACCACUGGCUACAGCUGGUUGAAGGGGACCAAAUGCUUGAAA-AA ((----((..((((....------------------)))).....)))).((((((((.(((.((.((((((((((....))).))))))).)).))).))).)))))...-.. ( -26.50) >DroSec_CAF1 123483 91 - 1 UG----GAAAAUAAUAAU------------------CCAUCGCAGUUCGCUAGGCUUUCGAUGUCAGCAACCACUGGCUACAGCUGGUUGAAGGGGACCAAAUGCUUAAAA-AA ((----((.........)------------------)))..(((((((.((.(((.......)))..(((((((((....))).)))))).)).))))....)))......-.. ( -22.70) >DroSim_CAF1 119437 91 - 1 UG----GAAAAUAAUAAU------------------CCAUCGCAGUUCCCUCGGCUUUCGAUGUCAGCAACCACUGGCUACAGCUGGUUGAAGGGGACCAAAUGCUUAAAA-AA ((----((.........)------------------)))..((((((((((.(((.......)))..(((((((((....))).)))))).)))))))....)))......-.. ( -27.10) >DroEre_CAF1 109799 105 - 1 -AGUUCACAGGUCACAGUUCACAGGUCACAGU-------GCACAGUUCACUCGAAUUUAGUUGGCAUCAACCACUGGCUACAGCUGGUUGAAGGGGACCAAAUGCUAGGAA-AA -..........((..(((.....((((....(-------((..(((((....)))))......)))((((((((((....))).)))))))....))))....)))..)).-.. ( -24.70) >DroYak_CAF1 113137 108 - 1 -A----AAAAAUAGUAAUUUAC-GCUUUCAGUCUGUCUAUCACAGUUUACUCGUACUUCGUUGUCAUCAACCACUGGCUACAGCUGGUUGAAGGGGACCAAAUGCUUGGAAAAA -.----................-..((((((.((((.....)))).......(((.((.(((.((.((((((((((....))).))))))).)).))).)).)))))))))... ( -23.80) >DroAna_CAF1 116920 80 - 1 ---------------AAU------------------AAAUCACUUUUCACUUGUACUUUGUUGUCAUCAACCACU-GCUACAGCUGGUUGAAGGGGACCAAAUGCUUGAAAAGA ---------------...------------------......(((((((...(((.((.(((.((.(((((((((-(...))).))))))).)).))).)).))).))))))). ( -24.00) >consensus _G____GAAAAUAAUAAU__________________CCAUCACAGUUCACUCGGAUUUCGUUGUCAUCAACCACUGGCUACAGCUGGUUGAAGGGGACCAAAUGCUUGAAA_AA ...........................................................(((.((.((((((((((....))).))))))).)).)))................ (-13.42 = -14.42 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:25 2006