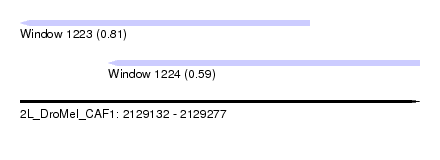
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,129,132 – 2,129,277 |
| Length | 145 |
| Max. P | 0.813041 |

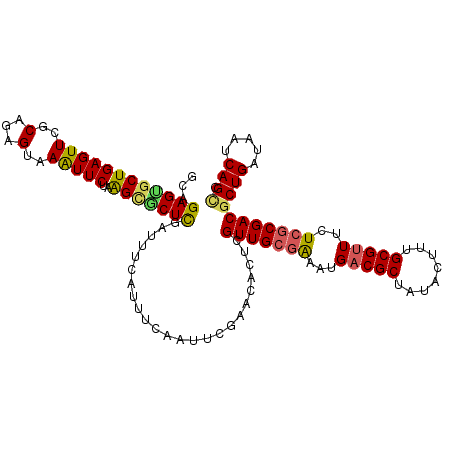
| Location | 2,129,132 – 2,129,237 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2129132 105 - 22407834 UUUCAUUUUGAUUCGAACACUCGUUGCGGAAUGUCGCUAUACUUUGCGUUUCUCACGACACUGAUAAUCAGCCUUGUGAAUAGUCAAUACUUUAACUACAAUCAG ...(((((((...(((....)))...))))))).(((........)))....((((((..(((.....)))..)))))).((((.((....)).))))....... ( -19.90) >DroSim_CAF1 128530 105 - 1 UUUCAUUUCGAUUCGAACACUCGUUGCGGAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCCUUGCGAGUGGUCAAAACUUUAACUACAAUCAG ..((((((((...(((....)))...))))))))(((........)))....((((((.((((.....)))).))))))(((((.((....)).)))))...... ( -32.30) >DroEre_CAF1 132092 105 - 1 UUUCAUUUCAUUUUGAACACUCGUUGCGAAAUGACGCUAUACCUUGCGUUUCUGGCGACGCUGAUAAUCAGUUUUGCGAAUUGCAAACACAAUAACAAAAAUCAG .......(((((((((((....))).)))))))).........(((((.(((..((((.((((.....)))).))))))).)))))................... ( -22.50) >DroYak_CAF1 134578 105 - 1 AUUCAUUUUAUUCGGAACACUCGUUGCGAAAUGACGCUUUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCUUUGCACUUUGUCAAAACUUAAACAACAAACAG ..............((.((...(((((((...(((((........)))))..)))))))((((.....)))).........)))).................... ( -23.60) >consensus UUUCAUUUCAAUUCGAACACUCGUUGCGAAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCCUUGCGAAUUGUCAAAACUUUAACAACAAUCAG ..((((((((...(((....)))...))))))))(((........)))....((((((.((((.....)))).)))))).......................... (-18.73 = -19.60 + 0.87)

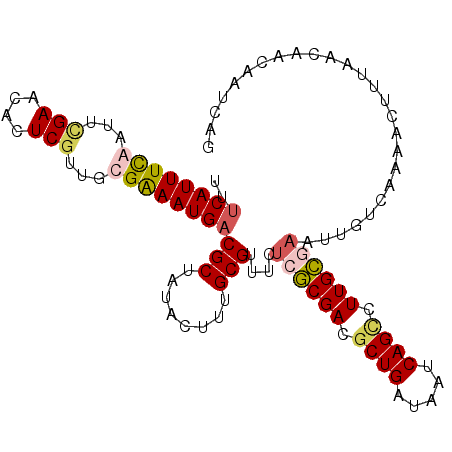
| Location | 2,129,164 – 2,129,277 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.73 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -26.90 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2129164 113 - 22407834 GCAGAGUGCUGAGUUCGCAGGGUAAGUUCUAAGUACUCGAUUUCAUUUUGAUUCGAACACUCGUUGCGGAAUGUCGCUAUACUUUGCGUUUCUCACGACACUGAUAAUCAGCC ....(((((((((..(((((((((.......(((..((((..((.....)).))))..)))....((((....))))..)))))))))...)))).).))))........... ( -29.10) >DroSim_CAF1 128562 113 - 1 GCAGAGCGCUGAGUUCGCCGGGUAAGUUCUAAGCGCUCGAUUUCAUUUCGAUUCGAACACUCGUUGCGGAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCC ((.(((((((.((..(.........)..)).)))))))....((((((((...(((....)))...)))))))).))......(((((.....))))).((((.....)))). ( -37.60) >DroEre_CAF1 132124 113 - 1 GCAGAGUGCUGAGUUCGCGGAGUAAAUUCUAGGCGCUUGAUUUCAUUUCAUUUUGAACACUCGUUGCGAAAUGACGCUAUACCUUGCGUUUCUGGCGACGCUGAUAAUCAGUU ...(((((..(((..(((..((......))..))))))...((((........)))))))))(((((.(...(((((........)))))..).)))))((((.....)))). ( -29.50) >DroYak_CAF1 134610 113 - 1 GCUGAGGGCUGAGUUCGCAGAGUAAAUUCUAAGAGCUCGGAUUCAUUUUAUUCGGAACACUCGUUGCGAAAUGACGCUUUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCU ((((((((((((((((..((((....))))..)))))))).))).......((((......((((((((...(((((........)))))..))))))))))))...))))). ( -42.10) >consensus GCAGAGUGCUGAGUUCGCAGAGUAAAUUCUAAGCGCUCGAUUUCAUUUCAAUUCGAACACUCGUUGCGAAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCC ...((((((((((((..(...)..)))))..)))))))........................(((((((...(((((........)))))..)))))))((((.....)))). (-26.90 = -27.02 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:32 2006