
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,595,051 – 20,595,211 |
| Length | 160 |
| Max. P | 0.907860 |

| Location | 20,595,051 – 20,595,171 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -29.68 |
| Energy contribution | -31.28 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.855030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20595051 120 + 22407834 AACUUGAUUGCCAUUUCAAUUGACCGCAGUUUUAGAAUGCCUUUUGCCAUAGUCAUGGGCAUCCAAAUGGGCUGUGGAAUUGGCAGCUACAGCUUUUGGCUUUUGUGAGACUGCGGCUGC ...(..((((......))))..)(((((((((((.((((((..((.((((((((.(((....)))....))))))))))..))))((((.......))))..)).))))))))))).... ( -42.80) >DroSec_CAF1 114546 120 + 1 AACUUGAUUGCCAUUUCAAUUGACCGCAGUUUUAGAAUGCCUUUUGCCAUGGUCAUGGGCAUCCAAAUGCGCUGUGGAAUUGGCAGCUACAGCUUUUAGCUUUUGUGAGACUGUGGCUGC ...(..((((......))))..)(((((((.((.(.((((((...((....))...)))))).).))...))))))).....(((((((((((((...((....))))).)))))))))) ( -40.30) >DroSim_CAF1 110321 120 + 1 AACUUGAUUGCCAUUUCAAUUGACCGCAGUUUUAGAAUGCCUUUUGCCAUGGUCAUGGGCAUCCAAAUGCGUUGUGGAAUUGGCAGCUACAGCUUUUGGCUUUUGUGAGACUGUGGCUGC ...(..((((......))))..)(((((((((((.((.(((...(((((...(((..(((((....)))).)..)))...)))))((....))....)))..)).))))))))))).... ( -39.80) >DroEre_CAF1 99777 108 + 1 AACUUGAUUGCCAUUUCAAUUGACCGCAGUUUUAGAAUGCCUCUUGCCAUAGUCAUGGGCACCCAAAUGG------------GCAGCUUCGGCUUUUGGCUUUUGUUGGGCUGCGGCUGC ...(..((((......))))..)(((((((((..(((.(((...((((...((..(((....))).)).)------------)))((....))....))).)))...))))))))).... ( -35.00) >DroYak_CAF1 103881 120 + 1 AACUUGAUUGCCAUUUCAAUUGACCGCAGUUUUAGAAUGCCUUUUGCAAUAGUCAUGGGAAUCCAAAUGGGGUGUGGAAUUGGCAGCUACAGCUUUUGGCUUUUGUUGGGCUGCGGAUGC ...(..((((......))))..)(((((((((..(((.(((...(((.....((((....((((.....)))))))).....)))((....))....))).)))...))))))))).... ( -31.40) >consensus AACUUGAUUGCCAUUUCAAUUGACCGCAGUUUUAGAAUGCCUUUUGCCAUAGUCAUGGGCAUCCAAAUGGGCUGUGGAAUUGGCAGCUACAGCUUUUGGCUUUUGUGAGACUGCGGCUGC ...(..((((......))))..)(((((((((((.(((((((...((....))...))))).(((((...(((((((.........))))))).)))))...)).))))))))))).... (-29.68 = -31.28 + 1.60)



| Location | 20,595,091 – 20,595,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -24.02 |
| Energy contribution | -25.82 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20595091 120 + 22407834 CUUUUGCCAUAGUCAUGGGCAUCCAAAUGGGCUGUGGAAUUGGCAGCUACAGCUUUUGGCUUUUGUGAGACUGCGGCUGCCUUUUGUCCACUUCCGCAUCAUUACGGACUCUGUCGCUUU .....((((.((((.(((....))).(((((..(((((...((((((..(((..(((.((....)).))))))..)))))).....)))))..)).))).......)))).))..))... ( -39.00) >DroSec_CAF1 114586 120 + 1 CUUUUGCCAUGGUCAUGGGCAUCCAAAUGCGCUGUGGAAUUGGCAGCUACAGCUUUUAGCUUUUGUGAGACUGUGGCUGCCUUUUGUCCACUUCCGCAUCAUUACGGACUCUGUCGCUUU .....((((.((((.(((....))).(((((..(((((...((((((((((((((...((....))))).))))))))))).....)))))...))))).......)))).))..))... ( -44.20) >DroSim_CAF1 110361 120 + 1 CUUUUGCCAUGGUCAUGGGCAUCCAAAUGCGUUGUGGAAUUGGCAGCUACAGCUUUUGGCUUUUGUGAGACUGUGGCUGCCUUUUGUCCACUUCCGCAUCAUUACGGACUCUGUCGCUUU .....((((.((((.(((....))).(((((..(((((...(((((((((((..(((.((....)).)))))))))))))).....)))))...))))).......)))).))..))... ( -44.40) >DroEre_CAF1 99817 94 + 1 CUCUUGCCAUAGUCAUGGGCACCCAAAUGG------------GCAGCUUCGGCUUUUGGCUUUUGUUGGGCUGCGGCUGCCUUUUGUCCACUUCCGCAUCAUUACG-------------- ....(((...(((...(((((.......((------------((((((.((((..(..((....))..))))).))))))))..))))))))...)))........-------------- ( -31.00) >DroYak_CAF1 103921 120 + 1 CUUUUGCAAUAGUCAUGGGAAUCCAAAUGGGGUGUGGAAUUGGCAGCUACAGCUUUUGGCUUUUGUUGGGCUGCGGAUGCCUUUUGUCCACUUCCGCAUCAUUACGGACUCCGUCGCUUU ..........(((.(((((..(((......(((((((((...((((((.((((...........))))))))))((((.......))))..))))))))).....))).))))).))).. ( -38.30) >consensus CUUUUGCCAUAGUCAUGGGCAUCCAAAUGGGCUGUGGAAUUGGCAGCUACAGCUUUUGGCUUUUGUGAGACUGCGGCUGCCUUUUGUCCACUUCCGCAUCAUUACGGACUCUGUCGCUUU ....(((........(((....)))...((...(((((...(((((((.((((((...((....))))).))).))))))).....)))))..)))))...................... (-24.02 = -25.82 + 1.80)


| Location | 20,595,091 – 20,595,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -24.75 |
| Energy contribution | -25.71 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
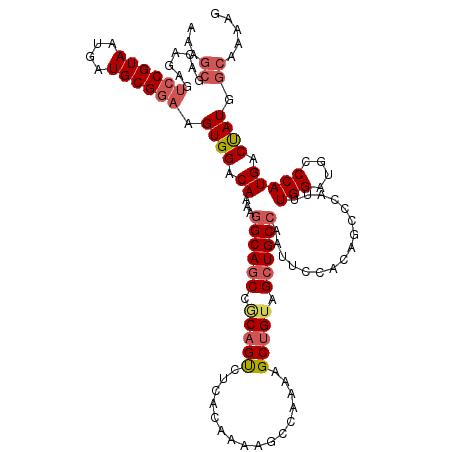
>2L_DroMel_CAF1 20595091 120 - 22407834 AAAGCGACAGAGUCCGUAAUGAUGCGGAAGUGGACAAAAGGCAGCCGCAGUCUCACAAAAGCCAAAAGCUGUAGCUGCCAAUUCCACAGCCCAUUUGGAUGCCCAUGACUAUGGCAAAAG ...((.......((((((....)))))).(((((.....((((((..((((................))))..))))))...))))).))((....)).((((.........)))).... ( -37.19) >DroSec_CAF1 114586 120 - 1 AAAGCGACAGAGUCCGUAAUGAUGCGGAAGUGGACAAAAGGCAGCCACAGUCUCACAAAAGCUAAAAGCUGUAGCUGCCAAUUCCACAGCGCAUUUGGAUGCCCAUGACCAUGGCAAAAG ...(((.(....((((((....)))))).(((((.....((((((.(((((................))))).))))))...))))).)))).......((((.((....)))))).... ( -38.99) >DroSim_CAF1 110361 120 - 1 AAAGCGACAGAGUCCGUAAUGAUGCGGAAGUGGACAAAAGGCAGCCACAGUCUCACAAAAGCCAAAAGCUGUAGCUGCCAAUUCCACAACGCAUUUGGAUGCCCAUGACCAUGGCAAAAG ...(((......((((((....)))))).(((((.....((((((.(((((................))))).))))))...)))))..))).......((((.((....)))))).... ( -39.09) >DroEre_CAF1 99817 94 - 1 --------------CGUAAUGAUGCGGAAGUGGACAAAAGGCAGCCGCAGCCCAACAAAAGCCAAAAGCCGAAGCUGC------------CCAUUUGGGUGCCCAUGACUAUGGCAAGAG --------------.....((.((.((..((((.(........)))))..)))).))...((((..(((....)))((------------((....))))...........))))..... ( -22.60) >DroYak_CAF1 103921 120 - 1 AAAGCGACGGAGUCCGUAAUGAUGCGGAAGUGGACAAAAGGCAUCCGCAGCCCAACAAAAGCCAAAAGCUGUAGCUGCCAAUUCCACACCCCAUUUGGAUUCCCAUGACUAUUGCAAAAG ...((((.((((((((....((((.((..(((((.....((((.(..((((................))))..).))))...)))))..))))))))))))))........))))..... ( -35.49) >consensus AAAGCGACAGAGUCCGUAAUGAUGCGGAAGUGGACAAAAGGCAGCCGCAGUCUCACAAAAGCCAAAAGCUGUAGCUGCCAAUUCCACAGCCCAUUUGGAUGCCCAUGACUAUGGCAAAAG ...((.......((((((....)))))).((((.((...((((((.(((((................))))).))))))................(((....))))).)))).))..... (-24.75 = -25.71 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:10 2006